OpenNeuro External Advisory Board Meeting 2023
Stanford University
Archive continues consistent growth
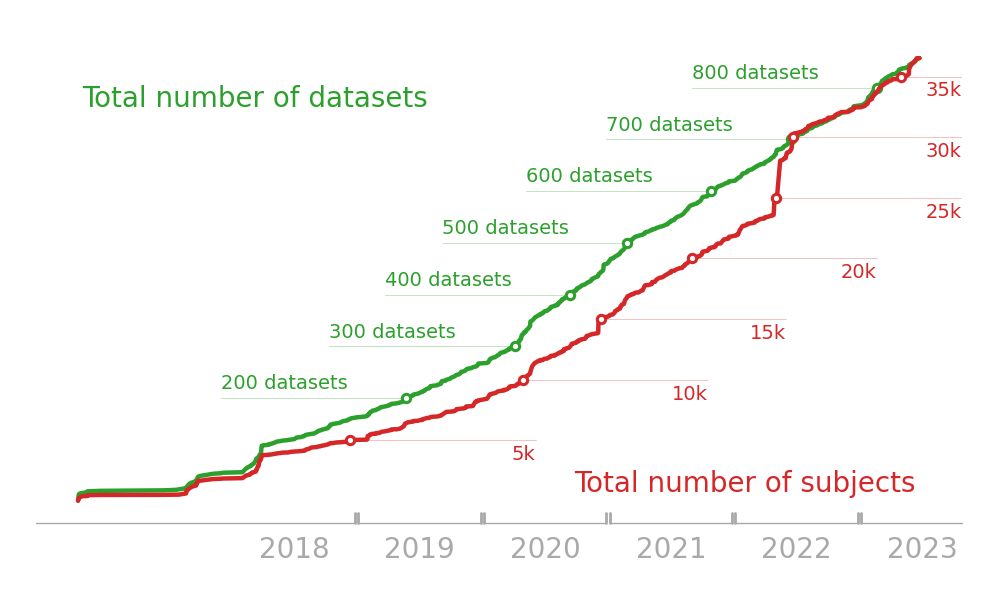
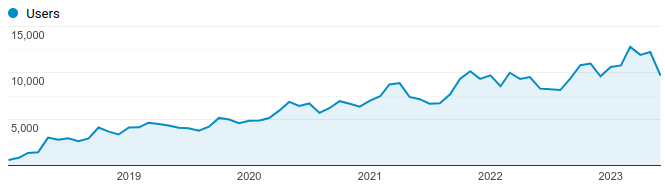
Increased web traffic over time
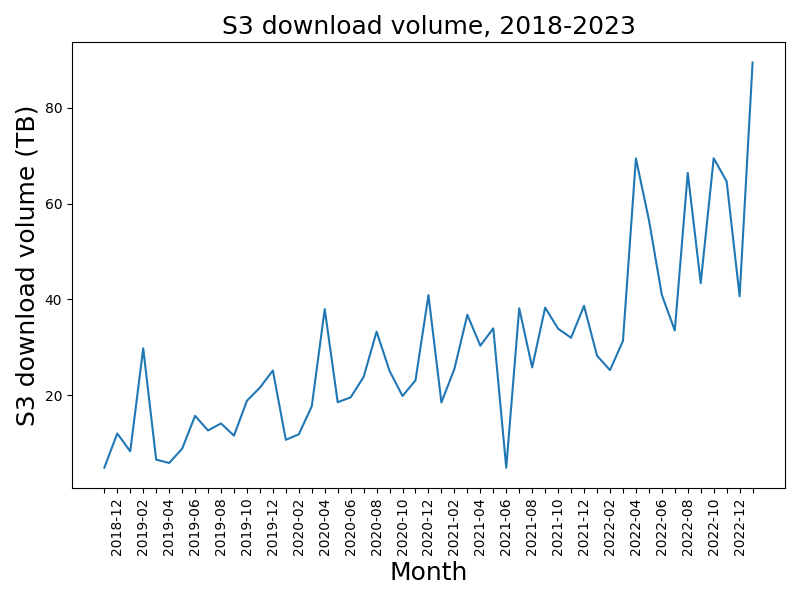
Substantial direct download volume
New programmatic developments
- NIH renewal grant approved for funding (2023-2028), awaiting NGA
- Foundation gift from the Noyce Foundation for Women’s Brain Health Initiative (2023-2025, w/ option for extension)
- Seed grant from Stanford Insitute for Human-Centered AI (with Sanmi Koyejo) on AI risks to privacy
- Planning to hire a full-time data curator (Joe Wexler, formerly part-time contractor)
- Considering hiring an additional software developer
New technical developments
- Migration from AWS to GCP
- Improved performance, easier scaling, and lower cost per dataset
- Open datasets still shared via AWS Public Datasets
- Improved performance for large datasets (now handling 2TB+)
- Published data retention/admin policies for reference in Data Sharing Plans
- Links to NEMAR for supported datasets
- Initial support for ORCID integration
- Support for fNIRS upload
Aims of renewal R24
- Overall goal
- Maintain the current high level of usability and performance of the OpenNeuro web site, through ongoing refinement of the site architecture
- Aim 1: Enhance utility for BRAIN Initiative projects
- Develop special landing page and support searching BRAIN Initiative datasets
- Allow custom data use agreements/sharing permissions for BRAIN Initiative investigators
- Aim 2: Enhance findability of OpenNeuro datasets
- Improve metadata access
- Aim 3: Enhance reusability of OpenNeuro datasets
- Support sharing of derivatives by users
- Provide standardized QA/preprocessed data
- Implement a Jupyterhub interface to OpenNeuro datasets (ala DANDI-hub)
Sharing of derivative datasets
- Derivative data (e.g. preprocessed MRI data) are now supported through the BIDS-Derivatives extension (developed under BRAIN R24MH114705)
- One major motivation for the schema-based validator was to support the validation of derivative datasets
- In advance of this, we have been developing support for sharing of derivative datasets within OpenNeuro
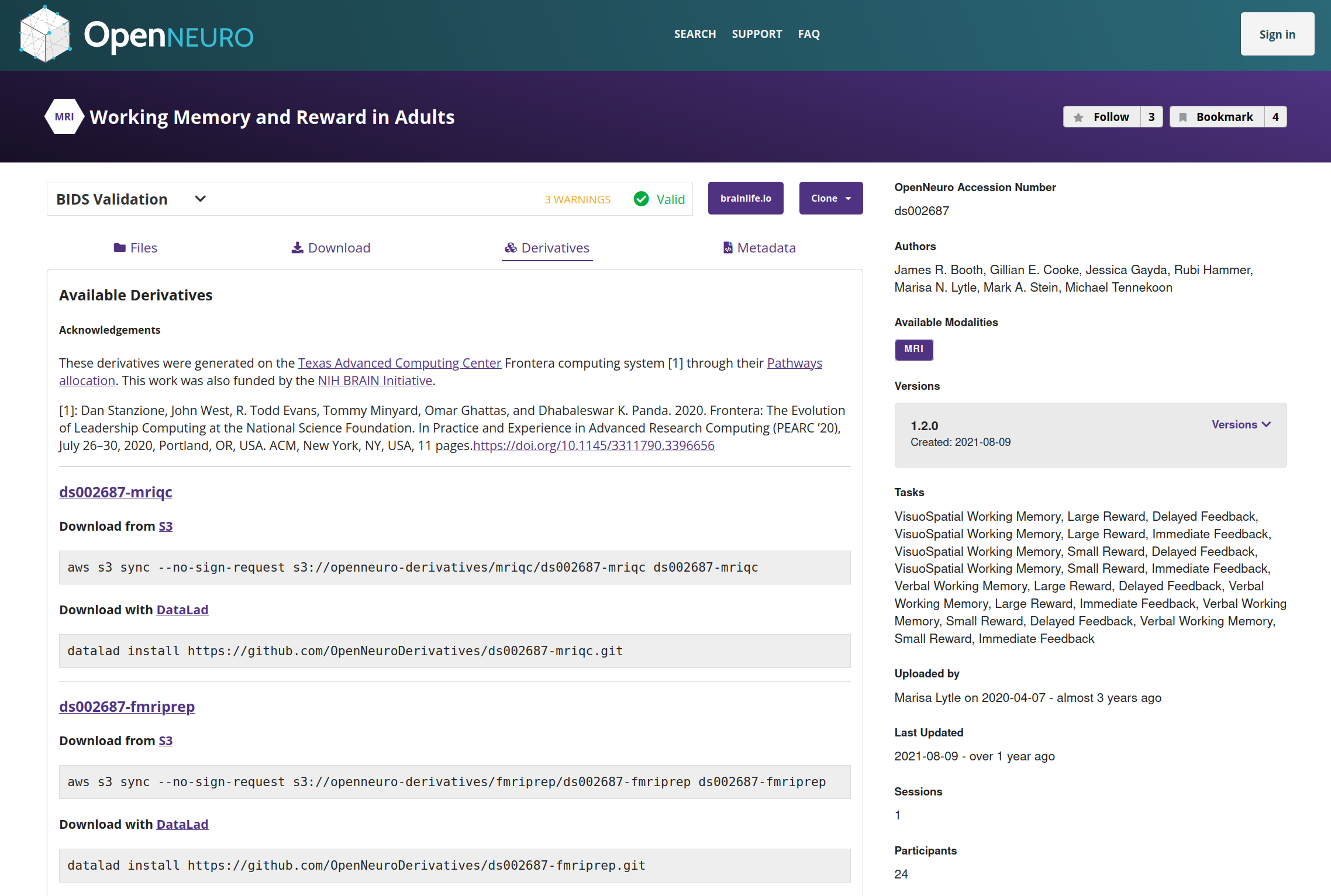
Preprocessing/QC of OpenNeuro fMRI datasets
- All human fMRI datasets from OpenNeuro are being run through MRIQC and fMRIPrep
- Via a Pathways allocation on the TACC Frontera supercomputer
- To date:
- 229/430 datasets successfully run with MRIQC
- 65/430 successfully preprocessed with fMRIPrep
- Derivatives openly available via S3 or Datalad
https://github.com/OpenNeuroDerivatives
Emerging issues
- Deidentification and privacy
- Hosting of clinical/higher-risk data
- Data use agreements
Alignment with new NIH Data Sharing Policy
“we highlight the importance of researchers considering whether, in choosing where and how to make their data available (if not already specified by an FOA or funding NIH ICO expectation), access to scientific data derived from humans should be controlled, even if de-identified and lacking explicit limitations on subsequent use…SACHRP identified concerns regarding re-identification of otherwise de-identified data, and indeed technological advances and increasing interoperability among data resources, while providing opportunities for new analyses, present identifiability concerns that are widely acknowledged.”
Final NIH Policy for Data Management and Sharing (https://grants.nih.gov/grants/guide/notice-files/NOT-OD-21-013.html)
Potential solution
- We are considering adding a click-through agreement for all users requiring agreement to not attempt reidentification
- This would cause friction because automated download would require an API key for each user
- vs. unauthenticated downloads from S3/datalad at present
- Open questions:
- Should we move to an authentication method that is more traceable to individuals (e.g. ORCID)? Or do we really want to be able to trace them?
- Does our agreement conflict with the CC0 licensing?
Neuroethics supplement
- A neuroethics supplement has supported Dr. Annie Jwa, a legal scholar with expertise in neuroethics
- Jwa & Poldrack (2022, Journal of Law and the Biosciences) argued for development of regulatory protections against misuse of neuroscience data
- “Neuroscience Information Non-discrimination Act”
- With seed grant funding from Stanford HAI, we are working with Sanmi Koyejo to examine potential for adversarial perturbations of structural MRI data to disable reidentification attempts

Hosting clinical data
- Should we tell users not to upload clinical data given the potentially higher risk to subjects from breach of privacy?
- We will be talking soon with ICPSR about potential use of OpenNeuro software for higher-risk data
The Poldrack Lab

Funding





OpenNeuro Team








Collaborators




Schema-based validation for BIDS
- OpenNeuro data ingestion relies upon JavaScript BIDS validator
- Original validator built the standard structure directly into the validator code
- Made addition of new data types quite laborious
- JavaScript expertise is relatively rare in our community
- Work began in 2021 on defining the standard separately using a schema that
- Three sprints to date involving the OpenNeuro team, NIMH Data Science and Sharing team, and others
Goals of the schema-based validator effort
- Authoritative, machine-readable descriptions of BIDS concepts
- Reduce need for proliferation of implementations, such as the PyBIDS configuration object
- Enforce consistency in specification by generating text and tables from schema
- Unify terms reused in multiple locations in the specification
- Reduce burden of writing BEPs by eliminating the requirement for validator coding
- Consequence: Whether a rule can be encoded in the schema or will need custom code / schema expansion is now an informal review criterion.
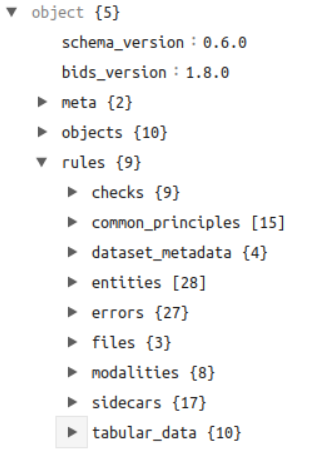
What is the schema?
- A hierarchy of YAML documents in the specification repository, under src/schema.
- Three major divisions:
- Objects (objects.*)
- Definitions of BIDS concepts like entities and terms like sidecar values
- Rules (rules.*)
- Validatable rules, such as entity ordering or permissible/required sidecar values
- Meta-schema (meta.*)
- Defines a “context” object to which rules can be applied
- Potentially expanded to any definitions or rules related to the schema itself
- Objects (objects.*)
Integrating the schema-based validator into OpenNeuro
- Completed
- Schema based validation can be run server side by OpenNeuro’s dataset worker
- Remaining
- Integration for client side (prior to upload) usage
- Port OpenNeuro CLI to new JavaScript runtime (Deno) to support calling the schema validator
- UI to control use of schema validator during upload
- Support for dual validation with existing and schema validators
- Decisions
- How do we handle cases when one validator passes and the other fails (for early users)?
https://poldrack.github.io/talks-openneuro/