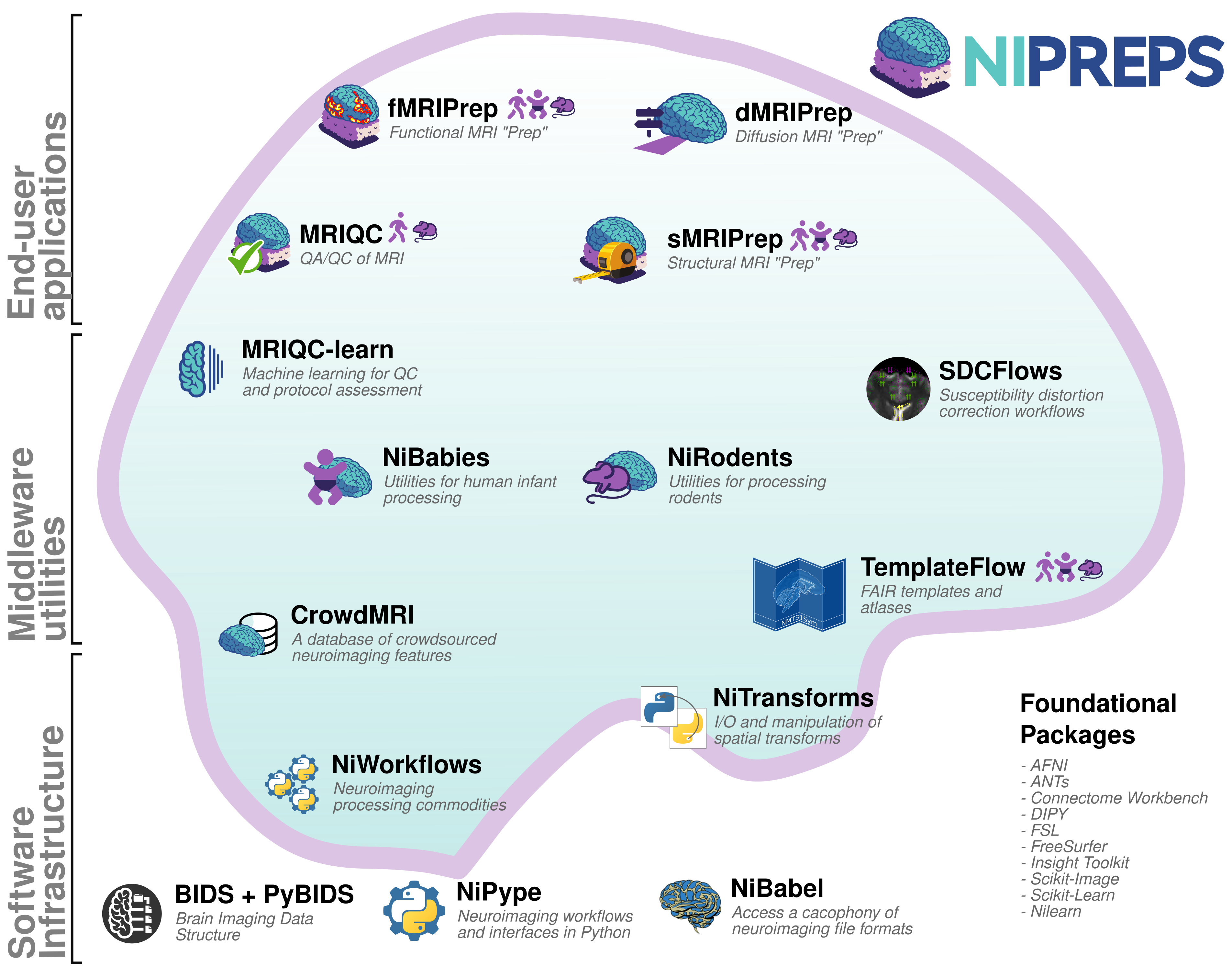
The NiPreps ecosystem for reproducible neuroimaging
Stanford University
The standard design (anti-)pattern for fMRI preprocessing
- Pick a single software package
- Usually based on considerations other than performance



The standard design (anti-)pattern for fMRI preprocessing
- Pick a single software package
- Usually based on considerations other than performance
- String together the tools from that package into a script to run the preprocessing workflow

The standard design (anti-)pattern for fMRI preprocessing
- Pick a single software package
- Usually based on considerations other than performance
- String together the tools from that package into a script to run the preprocessing workflow
- Generallly written by a student or postdoc with little software engineering experience
- Hope it keeps working over time…
- Trust that it does the right thing…

Towards a new design pattern for preprocessing: fMRIPrep
- A robust workflow for preprocessing fMRI data
- Tested on a large number of fMRI datasets from OpenNeuro
- Adapt to each dataset and processes it in the best way possible
- Outputs to BIDS-Derivatives format
- Provides powerful visualizations :
- to help identify problems
- allowing glass-box access to training researchers
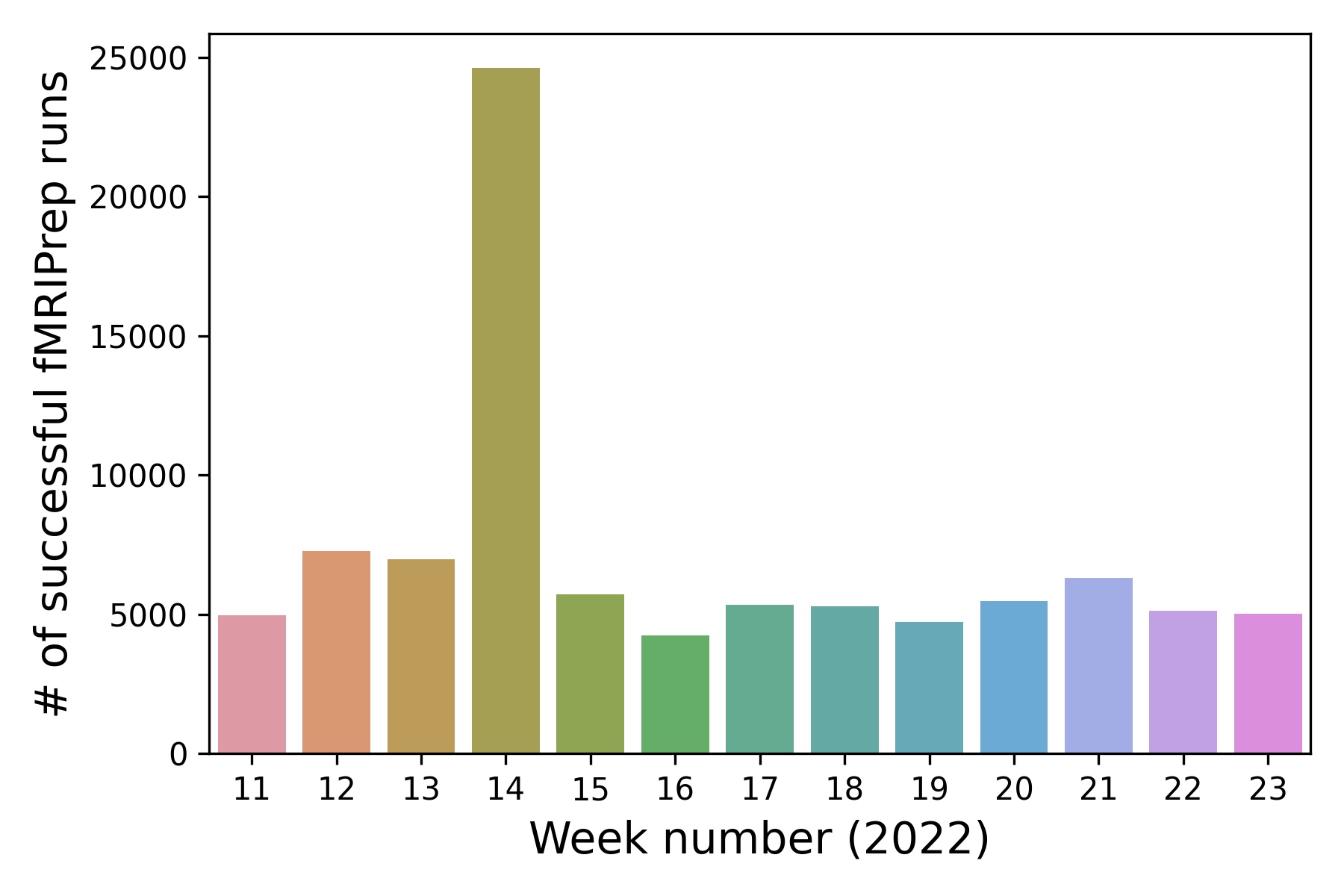
- Currently averaging ~5000 successful runs per week
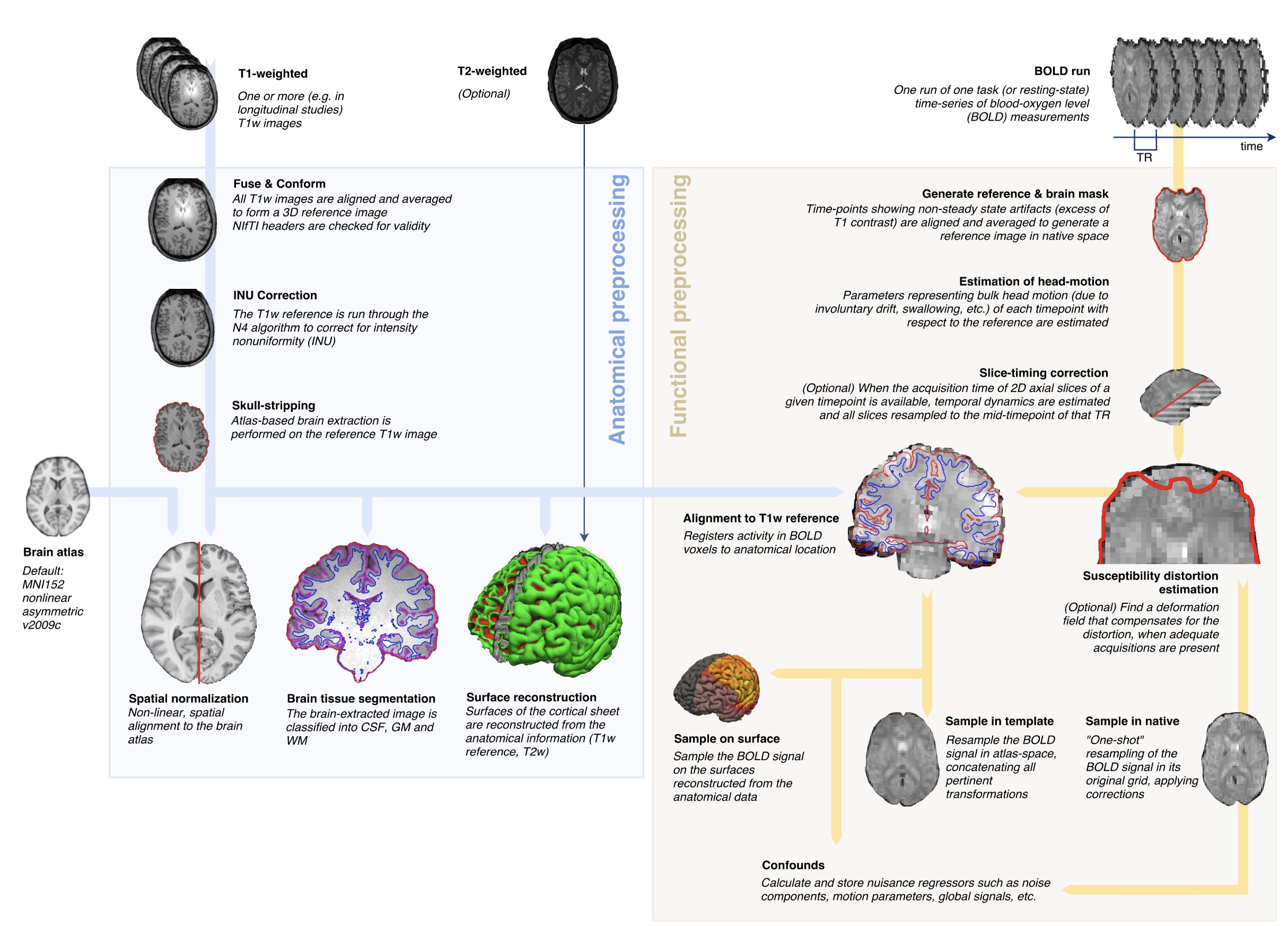 Esteban et al., 2019, Nature Methods
Esteban et al., 2019, Nature Methods
RF1MH121867: NiPreps (NeuroImaging PREProcessing toolS)
… the overarching objective of this project is to develop NiPreps, a software framework to perform standardized preprocessing of diverse neuroimaging data.
- Aim 1: solidify the foundations of the NiPreps integration.
- Aim 2: enable integrative analysis approaches of heterogeneous data.
- Aim 3: accelerate the dissemination of NiPreps to the neuroscience community through hackathons and “docusprints”.
Why?
In developing fMRIPrep, we learned about critical aspects of neuroimaging workflows. NiPreps is envisioned as a generalization of fMRIPrep.
RF1MH121867: Sites

Poldracklab (Stanford)


Esteban Lab (CHUV Lausanne)

Milham Lab (Child Mind Inst)
Rokem Lab (UW)
The NiPreps community
www.nipreps.org
Aim 1: Developing and refining reusable infrastructure/middleware components
- TemplateFlow: FAIR Sharing and management of neuroimaging templates and atlases
- SDCflows: Integrating susceptibility distortion correction (SDC)
- NiReports: A modular visual reports system
- NiTransforms: Spatial transforms integration
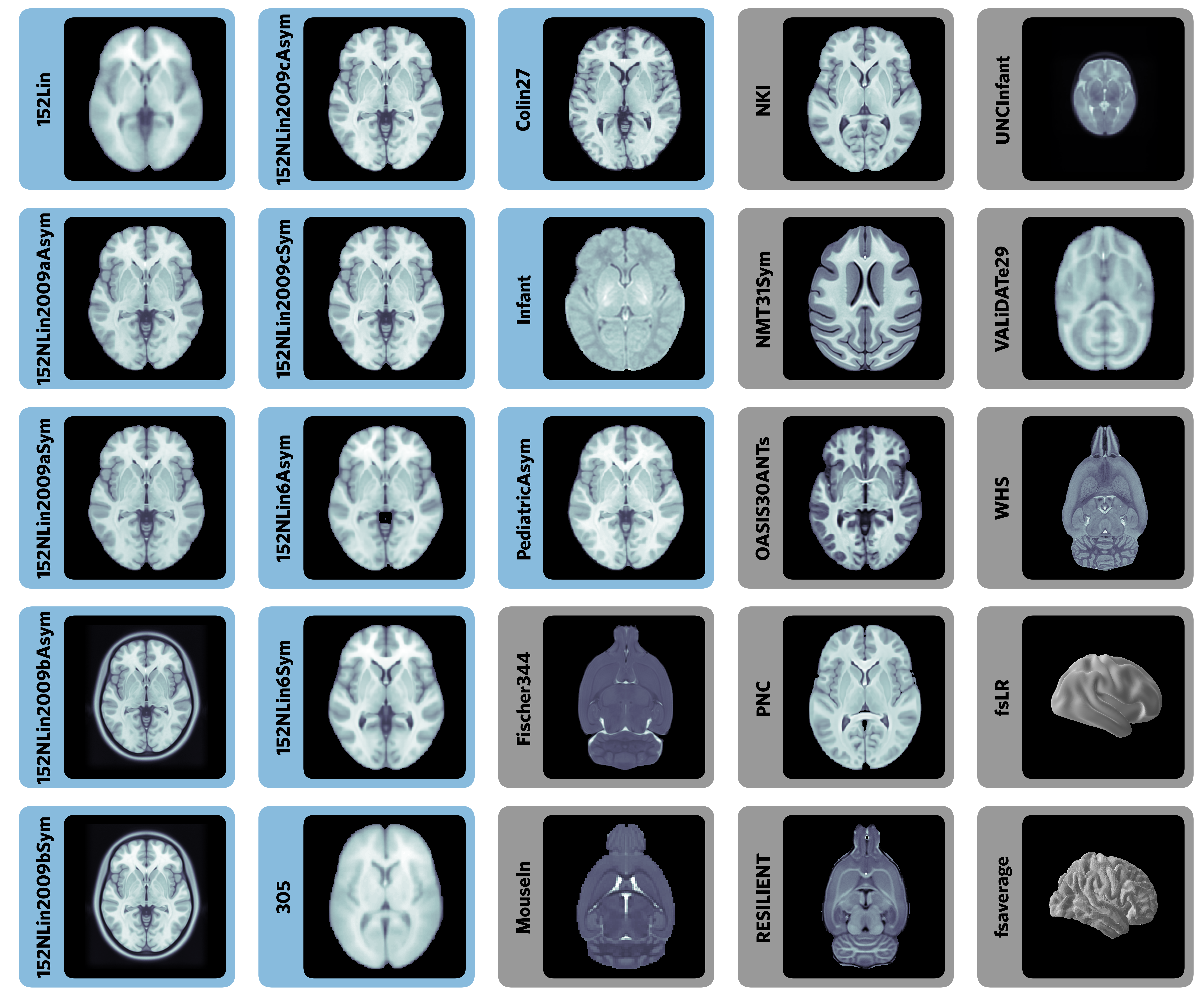
TemplateFlow: FAIR sharing of neuroimaging templates and atlases
Templates and atlases are commonly used in neuroimaging research
There is significant lack of clarity in the use of these templates
- There are numerous versions of the widely used “MNI template”
Templateflow provides programmatic access to a database of templates and mappings between them
Easy to use for humans and machines:

 Ciric et al., 2022, Nature Methods
Ciric et al., 2022, Nature Methods
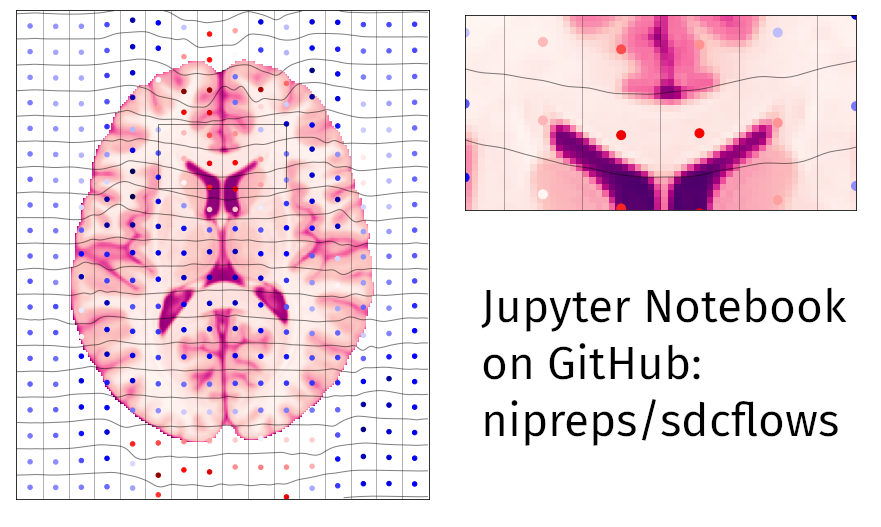
SDCFlows: Susceptibility Distortion Correction workflows
- SDCflows aims to provide a unified interface to susceptibility distortion correction methods
- Defines a shared representation model (B-Spline) for the field map
- “decouples” estimation and application steps (increasing modularity)
- Overhaul started early 2021 (Esteban et al., OHBM 2021)
- Faced many technical challenges
- Requiring numerous bugfixes and “edge” cases
- Developed new educational materials & Jupyter notebooks
- Faced many technical challenges
NiReports: New visualization tools from MRIQC
- MRIQC is a quality control workflow for structural/functional MRI
- Developing a number of visualizations that will go into NiReports
- Added visualization of voxels at the edge of the brain (“crown”)
- Added hierarchical sorting of rows (voxels) to enhance patterns (Aquino et al. 2019)
Infrastructure: Architectural redesign
- Problem: fMRIPrep’s “one size fits all” design has limitations for emerging use cases
- Archiving preprocessing results requires balancing storage costs against possible use cases.
- Including alternative algorithms requires custom code to integrate.
- Solution: Accept pre-computed derivatives and defer computationally cheap operations
- E.g., Deep learning segmentations and masks can be accepted, skipping fMRIPrep defaults.
- Multiple template registrations can be archived, analysts may resample BOLD series to different spaces on demand.
- This approach is implemented in SDCFlows and is being generalized to other components.
Aim 2: Expand the portfolio of end-user NiPreps
- ASLPrep
- dMRIPrep
- PETPrep
- fMRIPrep-infants (aka NiBabies)
- fMRIPrep-rodents (aka NiRodents)
Workflows: ASLPrep (cerebral blood flow quantification)
- A robust workflow for preprocessing arterial spin labeling (ASL) data
- Including cerebral blood flow (CBF) quantification
- Provides quality evaluation for CBF maps
- Provides CBF quantification at the regional level using atlases
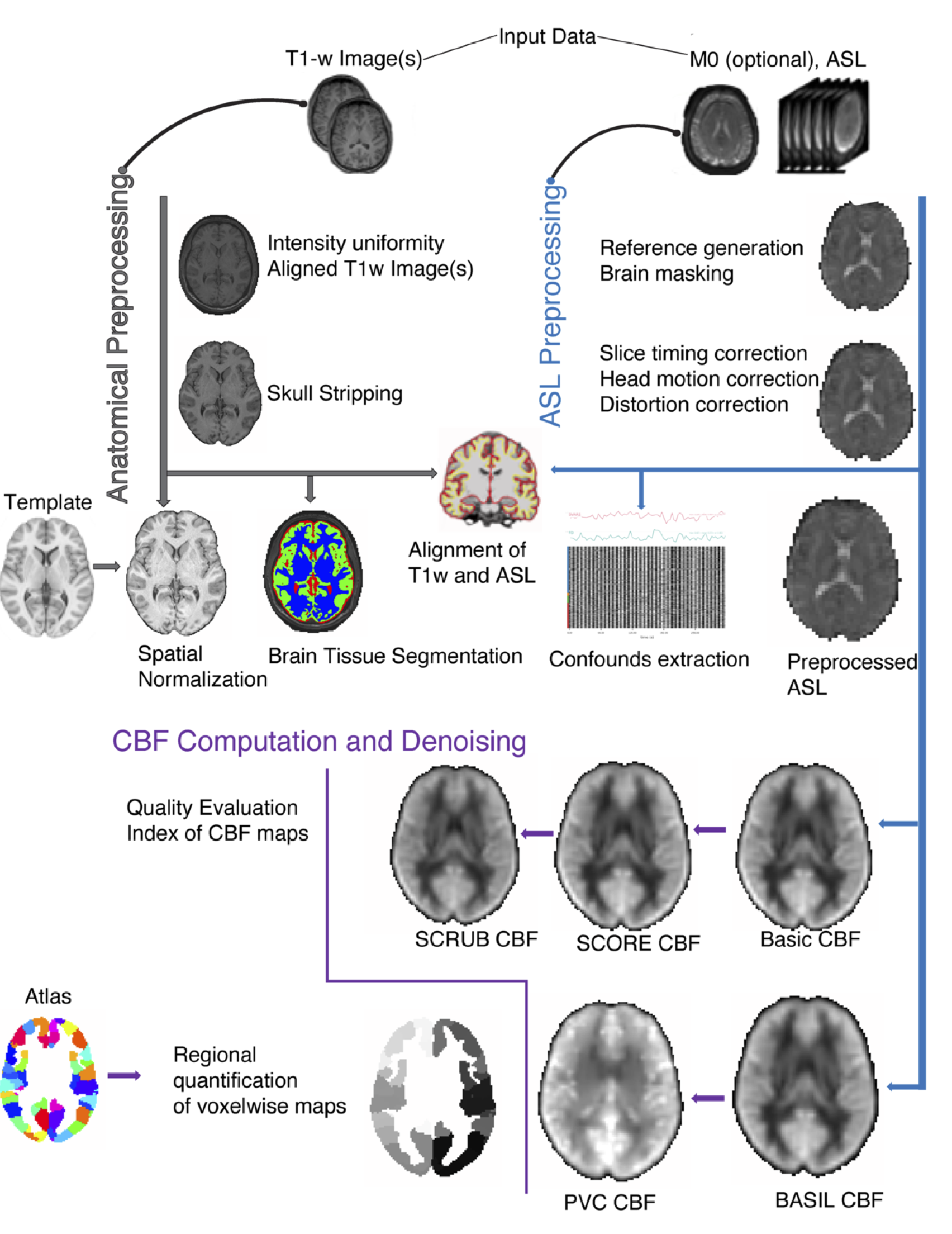
Adepimbe et al., 2022, Nature Methods
Workflows: dMRIPrep (diffusion MRI)
- A workflow for preprocessing of diffusion MRI data
- Development currently focused on eddymotion
- an algorithm to estimate head-motion (modality-agnostic) and modality-specific artifacts (eddy currents in the case of dMRI)
Preprint: Pisner et al., 2022.
Workflows: PETPrep (positron emission tomography)
- A NiPreps workflow for PET preprocessing
- Successfully merged petsurfer into nipype (1.8.0)
- Incorporated nipype implementation of a robust head motion correction workflow (petprep_hmc)
- Developing a BIDS-Derivatives standard for PET derivatives
- Model-based head-motion correction leveraging eddymotion in progress
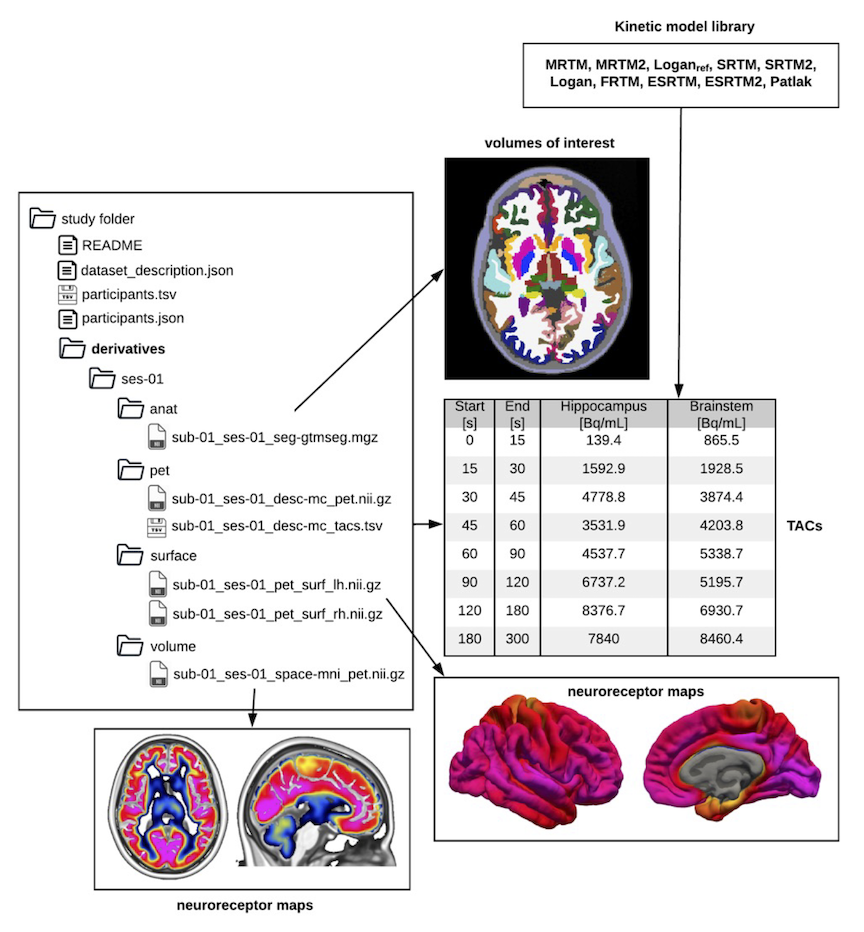
Martin Norgaard, in prep
Workflows: fMRIPrep for infants
- Collaboraration with Damien Fair & HBCD team
- New developments
- Support for pre-computed derivatives (mask, segmentations).
- Improved robustness and validity of CIFTI-2 outputs.
- Upcoming developments
- Morphometric outputs (cortical thickness, curvature)
- Improvements to susceptibility distortion correction versatility
- T2 assisted surface generation
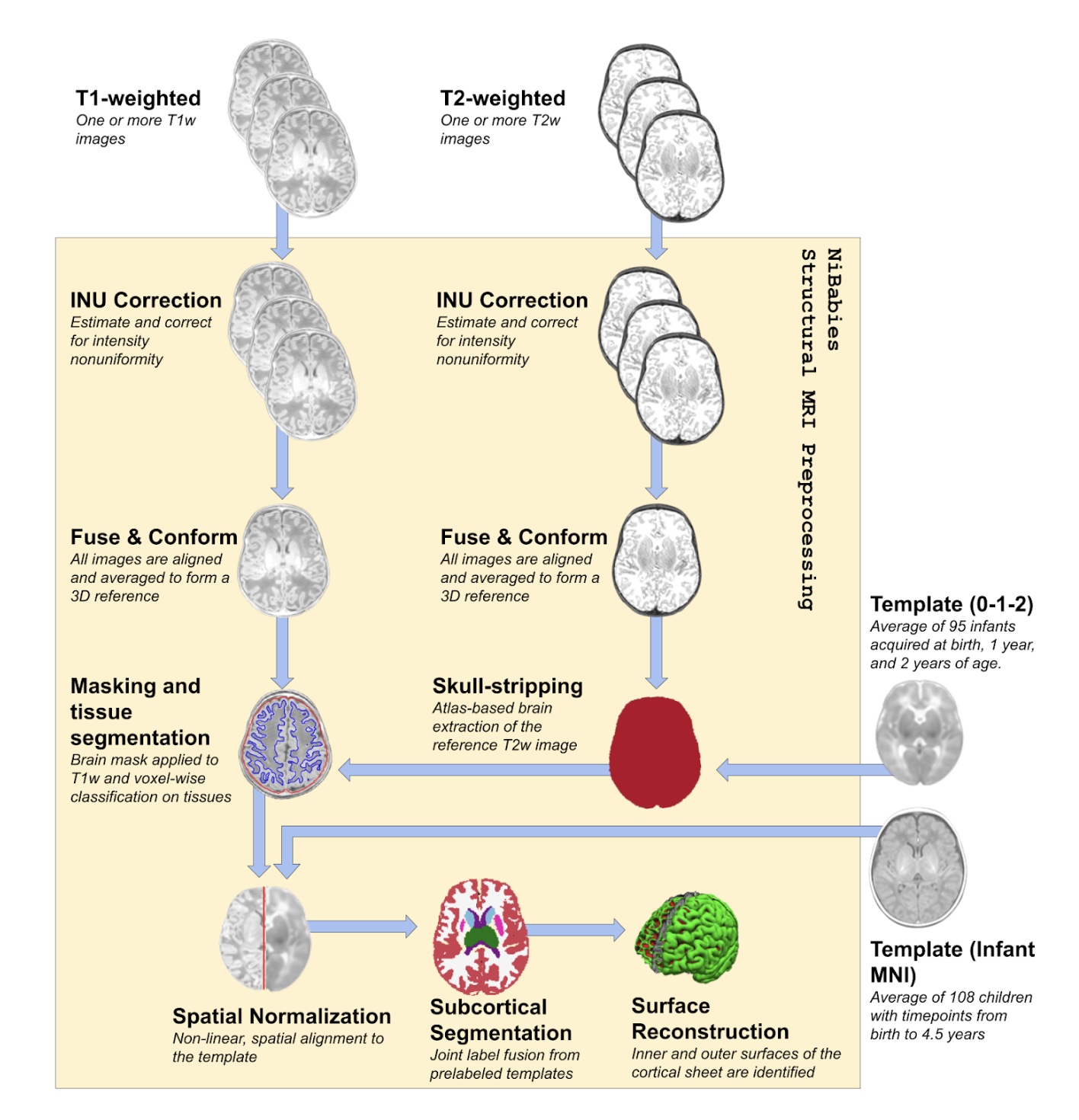
Mathias Goncalves, in prep
Aim 3: Consolidate the NiPreps community
- Project monitoring infrastructure: MIGAS
- Evaluation of cross-workflow reproducibility
- Hackathons and documentation
- Best practices and educational resources
- NMIND: Building common standards for software development
Project monitoring: MIGAS
- An open-source, customizable telemetry solution
- Allows collecting usage information, errors, and status throughout a process’s lifetime
- Easy to deploy with various cloud providers (Heroku / GCP / AWS)
- Available as a Python package: https://pypi.org/project/migas/
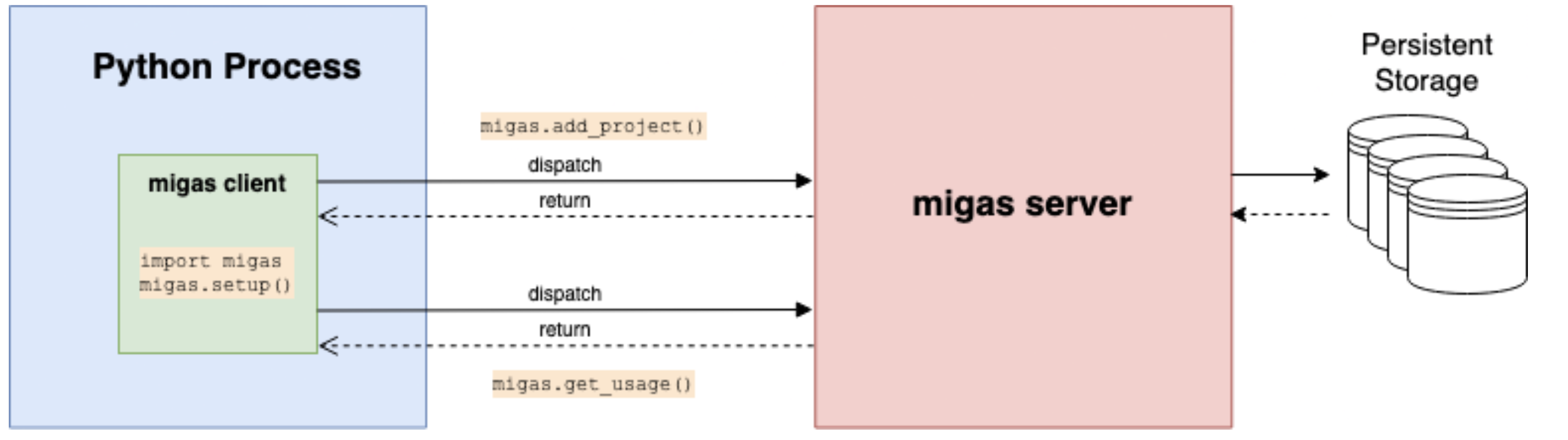
Mathias Goncalves, in prep
Reproducibility: Cross-workflow evaluation
- CMI team developed a CPAC implementation of fMRIPrep
- Able to achieve high levels of reproducibility in connectivity metrics between harmonized workflows
- Helped identify causes of divergence, such as use of different versions of MNI template
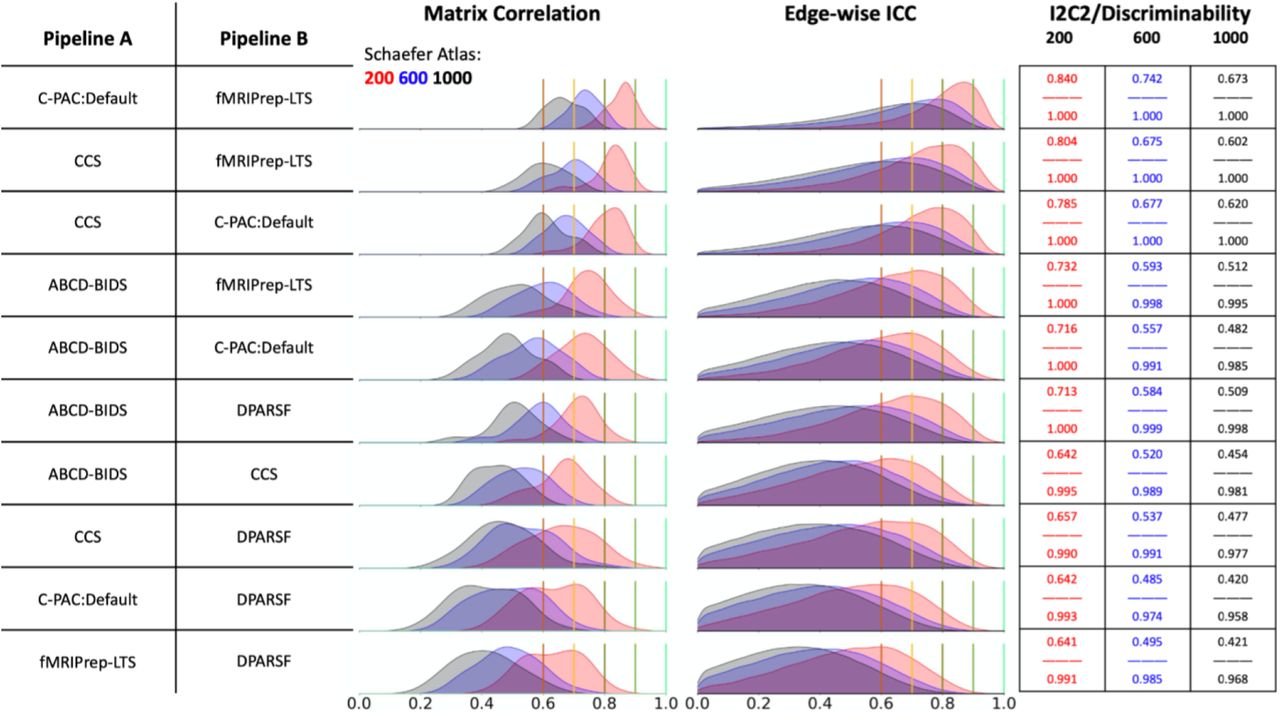
Xinhui Li et al., under review
Hackathons and documentation
- Held a hackathon/documentation sprint in Glasgow following OHBM 2022
- Will participate in Brainhack Global 2022
- Planning to hold a hackathon/documentation sprint in Montreal in association with OHBM 2023
Best practices and educational resources
Collaborative QC-Book (educational, ISMRM 2021): https://nipreps.org/qc-book
MRIQC-SOPs (standard operating procedures)
- A GitHub template-repository to create and maintain versioned SOPs documentation and checklists.
- Example: https://nipreps.org/mriqc-sops/
MRIQC Protocol report (Hagen et al., in preparation)
Frontiers’ research topic on QC of fMRI (Provins et al., under review)
Biases introduced by defacing in QC (Provins et al., pre-registered report under review)
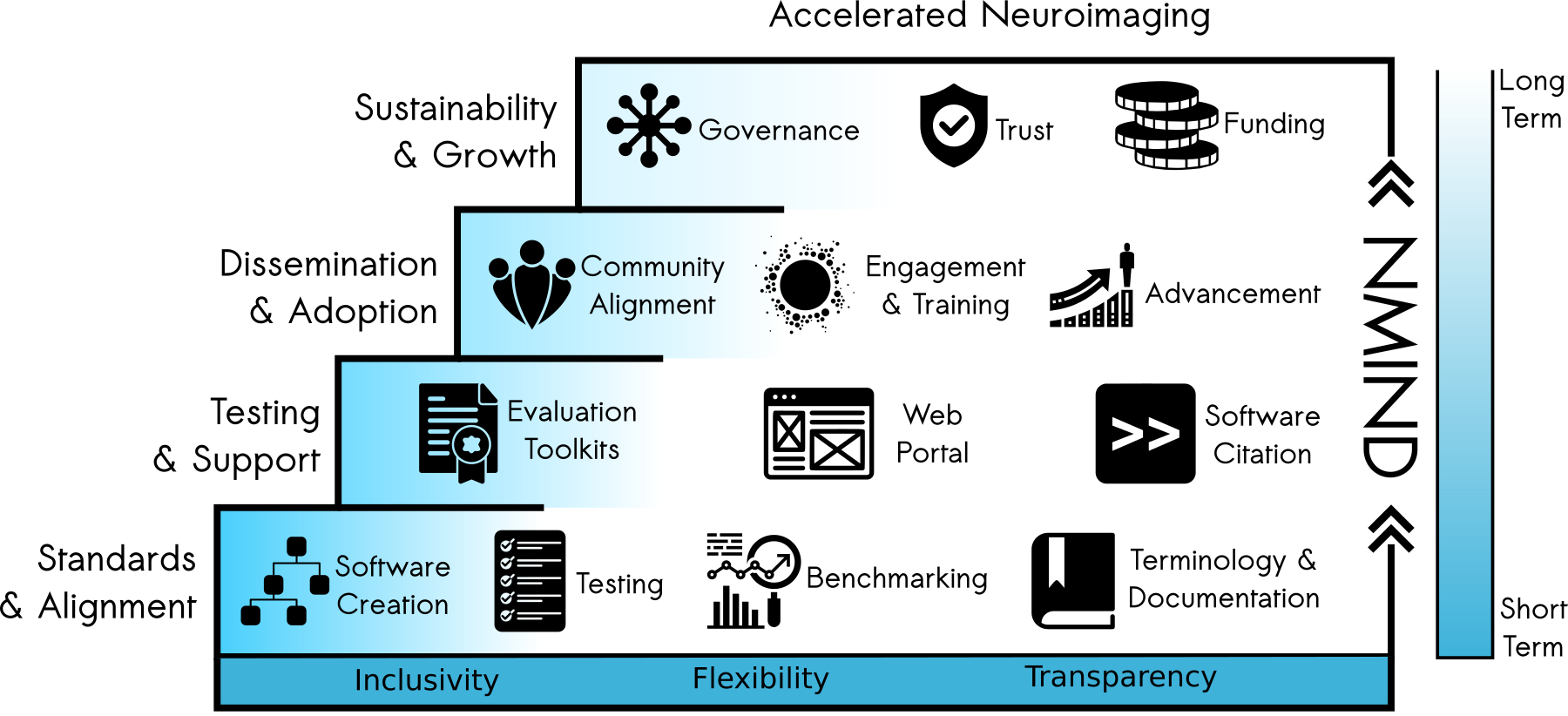
NMIND: Building common standards for software development
- NMIND: Nevermind, this Method Is Not Duplicated
- Alignment: development and adoption of standards for critical software component
- Testing: accessible and (semi-)automated mechanisms for evaluating standards compliance
- Engagement: widespread promotion and adoption of the NMIND collaborative standards
Thank you!
fMRIPrep usage
- Usage tracked using an opt-out telemetry system
- Allows quick identification of bugs and usage patterns
- Currently averaging ~5000 successful runs per week
Note
We are developing an open-source alternative called “migas”, to replace Sentry
SDCFlows in a nutshell
- What? Wraps or implements methods for estimating field maps in various scenarios:
- covering (i) so-called TOPUP, (ii) phase-difference fieldmaps and their variants, (iii) fMRIPrep’s “fieldmapless”; but
- it does not cover point-spread-function approaches (extremely marginal).
- How?
- Defines a shared representation model (B-Spline) for the field map (comparability ↑↑, methodological variability ↓↓)
- Technical perk: “decouples” estimation and application steps (modularity ↑↑, methodological variability ↑).
- Why?
- A single tool can be applied to correct for distortion, no matter how it was estimated (method comparability ↑↑)
- The model coefficients can easily be moved with head motion (e.g., dynamic fieldmap)
- The model coefficients can easily be integrated in spatial transformation chains (one-shot interpolation)
- The model coefficients can easily be integrated within other software (e.g., eddymotion)
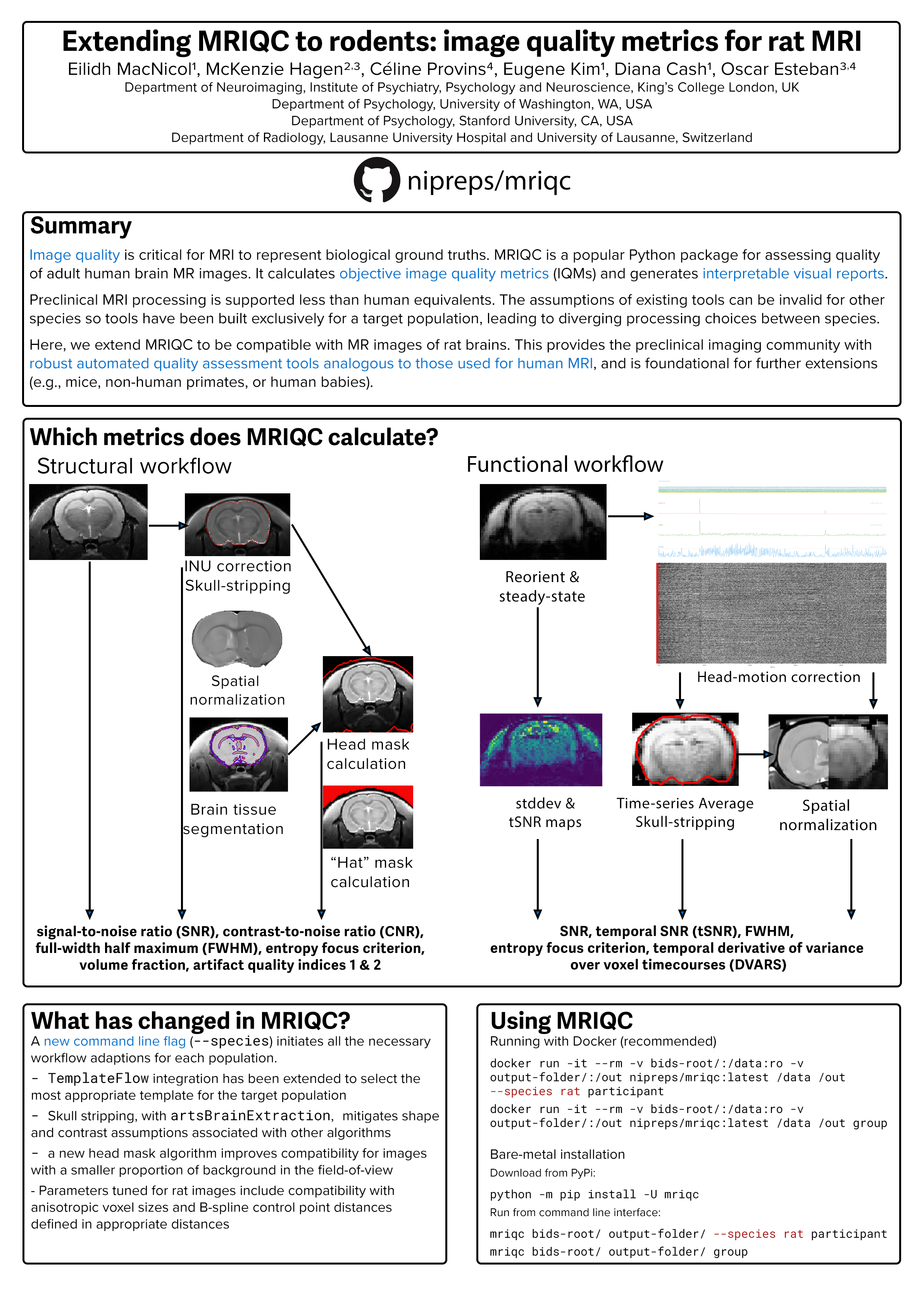
Result YR1: NiRodents
- MRIQC-rodents saw a first release presented in EMIM 2022
- fMRIPrep-rodents is currently integrating new SDCFlows’ API
- NiRodents has stimulated several improvements of the reporting system (see next)
- NiRodents has stimulated the inclusion (and revision of existing) rodent templates in TemplateFlow
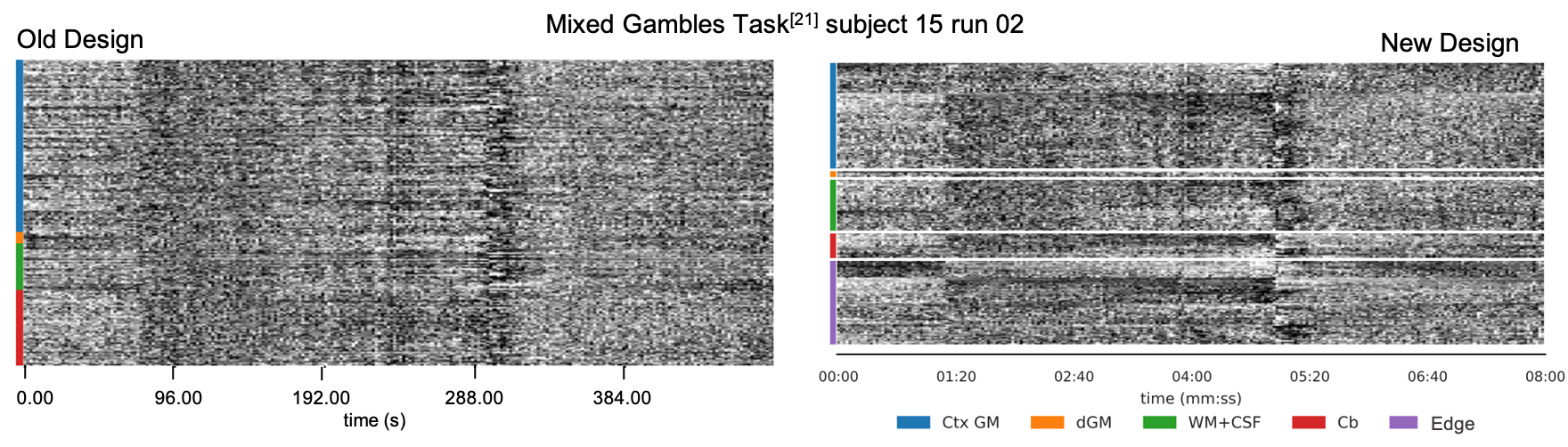
Result YR1: new visualizations in MRIQC and fMRIPrep
- Added visualization of voxels at the edge of the brain (“crown”)
- Added hierarchical sorting of rows (voxels) to enhance patterns (Aquino et al. 2019)
- fMRIPrep: added “edge” regressors (Patriat et al. 2008)
Results YR1: Other areas
- NiReports: provides standard mechanisms to build “reportlets” and full reports.
- A repository has been initiated
- Code already exists, but it is scattered across tools (e.g., MRIQC, fMRIPrep)
- In the OHBM Hackathon, work initiated for moving MRIQC visual components into it (fMRIPrep will follow).
- NiTransforms:
- The component is in a stable status
- The component is key to achieving a redesign of fMRIPrep (described next)
https://poldrack.github.io/talks-nipreps/
 (
(