Toward an Open Science Ecosystem in Neuroimaging
Stanford University
Transparency is essential for reproducibility

“we can distill Claerbout’s insight into a slogan:
An article about computational science in a scientific publication is not the scholarship itself, it is merely advertising of the scholarship. The actual scholarship is the complete software development environment and the complete set of instructions which generated the figures..”
- Buckheit & Donoho, 1995

Jon Claerbout
Why neuroimaging is a best-case scenario for open science
- Magnetic resonance imaging (MRI) is the primary tool for studying human brain structure and function
- MRI data are digital end-to-end
- From MRI scanner to automated analysis
- Usually zero/few manual analysis steps
- The field has largely converged on:
- a standardized image format (NiFTI)
- a ~common spatial coordinate system
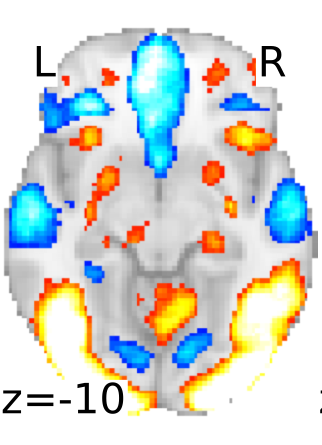
A false start for fMRI data sharing

A false start for fMRI data sharing


This letter comes from a group of scientists who are publishing papers using fMRI to understand the links between brain and behavior. We are writing in reaction the recent announcement of the creation of the National fMRI Data Center (www.fmridc.org). In the letter announcing the creation of the center, it was also implied that leading journals in our field may require authors of all fMRI related papers accepted for publication to submit all experimental data pertaining to their paper to the Data Center. … We are particularly concerned with any journal’s decision to require all authors of all fMRI related papers accepted for publication to submit all experimental data pertaining to their paper to the Data Center.
2010: The year data sharing broke in neuroimaging

- “Comprehensive mapping of the functional connectome, and its subsequent exploitation to discern geneticinfluences and brain–behavior relationships, will require multicenter collaborative datasets. Here we initiate this endeavor by gathering R-fMRI data from 1,414 volunteers collected independently at 35 international centers. We demonstrate a universal architecture of positive and negative functional connections, as well as consistent loci of inter-individual variability. …”
Data sharing is becoming the norm in neuroimaging
Poldrack et al., Annual Reviews in Biomedical Data Science, 2019
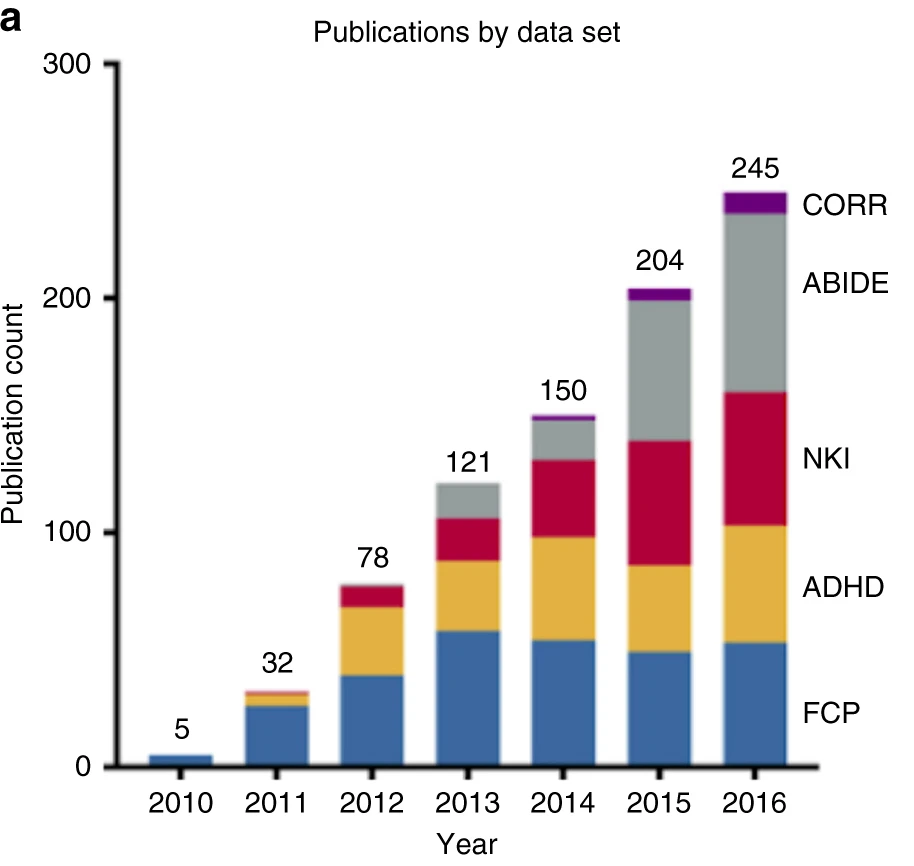
Milham et al., Nature Communications, 2018
Anonymous senior researcher:
“OHBM has been taken over by the open science zealots!”
An open ecosystem for retrospective data sharing
- Neurosynth.org: Open database of published neuroimaging coordinates
- Neurovault.org: Open archive for neuroimaging results
- OpenNeuro.org: Open archive for raw/processed neuroimaging data
Maximally open sharing
- Data shared under maximally permissive data use agreements:
- Neurosynth: Open Data Commons Open Database License v1.0
- Neurovault: CC0
- OpenNeuro: CC0
- All data available programmatically via web API as well as web page

- CC0 enables scientists, educators, artists and other creators and owners of copyright- or database-protected content to waive those interests in their works and thereby place them as completely as possible in the public domain, so that others may freely build upon, enhance and reuse the works for any purposes without restriction under copyright or database law.
- https://creativecommons.org/share-your-work/public-domain/cc0/
Neurosynth: Sharing activation coordinates
- Brain activity is reported in a (somewhat) standardized coordinate system
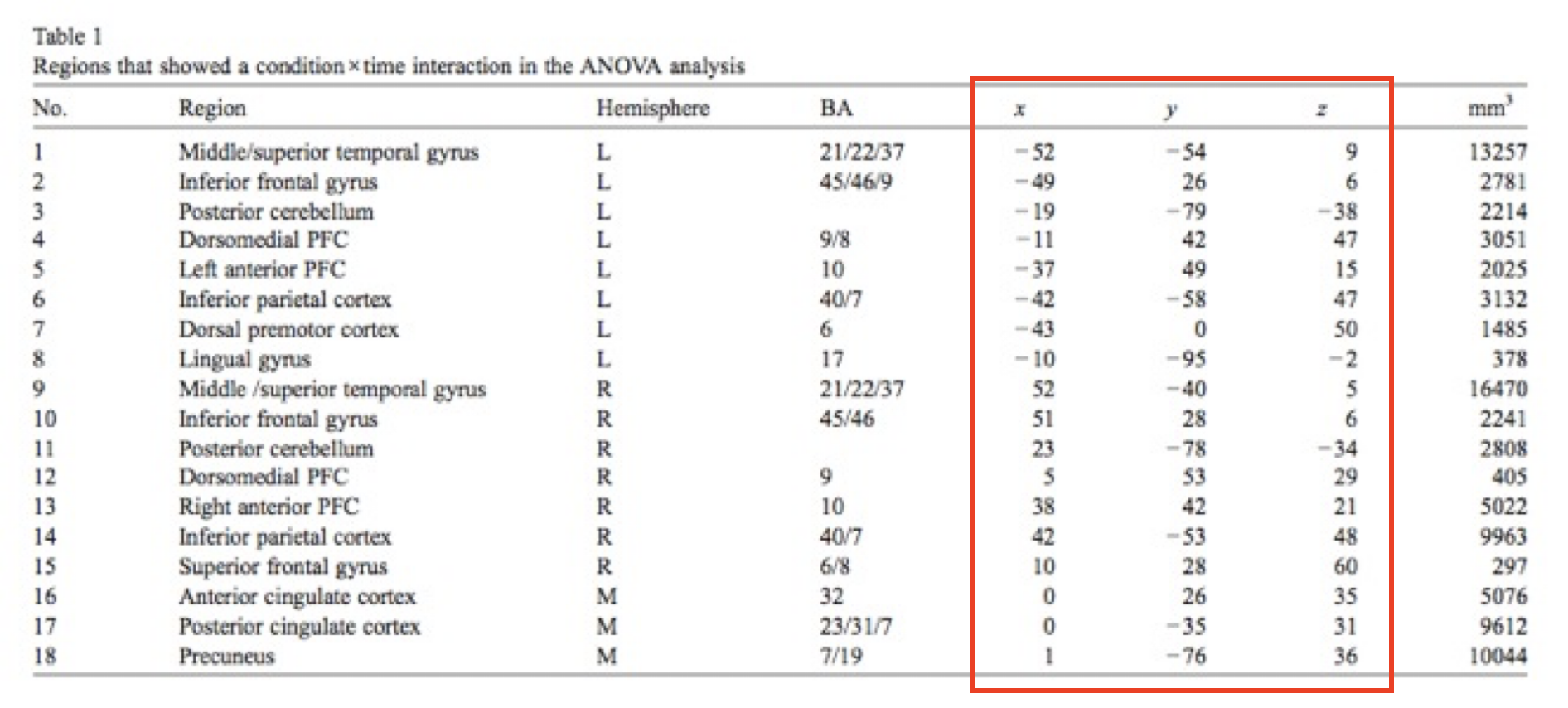

Creating meta-analytic maps
- Automated Coordinate Extraction
- Automatically extracts activation tables from fMRI papers for 17 journals
- Current database has 14,371 papers (with full text)
- 84% sensitivity, 97% specificity against manual database (SumsDB)
- Meta-analytic maps created for each paper
- 10mm sphere placed at each focus
| X | Y | Z |
|---|---|---|
| 12 | 57 | -6 |
| 33 | 21 | 15 |
| 24 | -6 | 51 |
| 28 | 10 | 18 |
↓
Yarkoni et al, 2011, Nature Methods
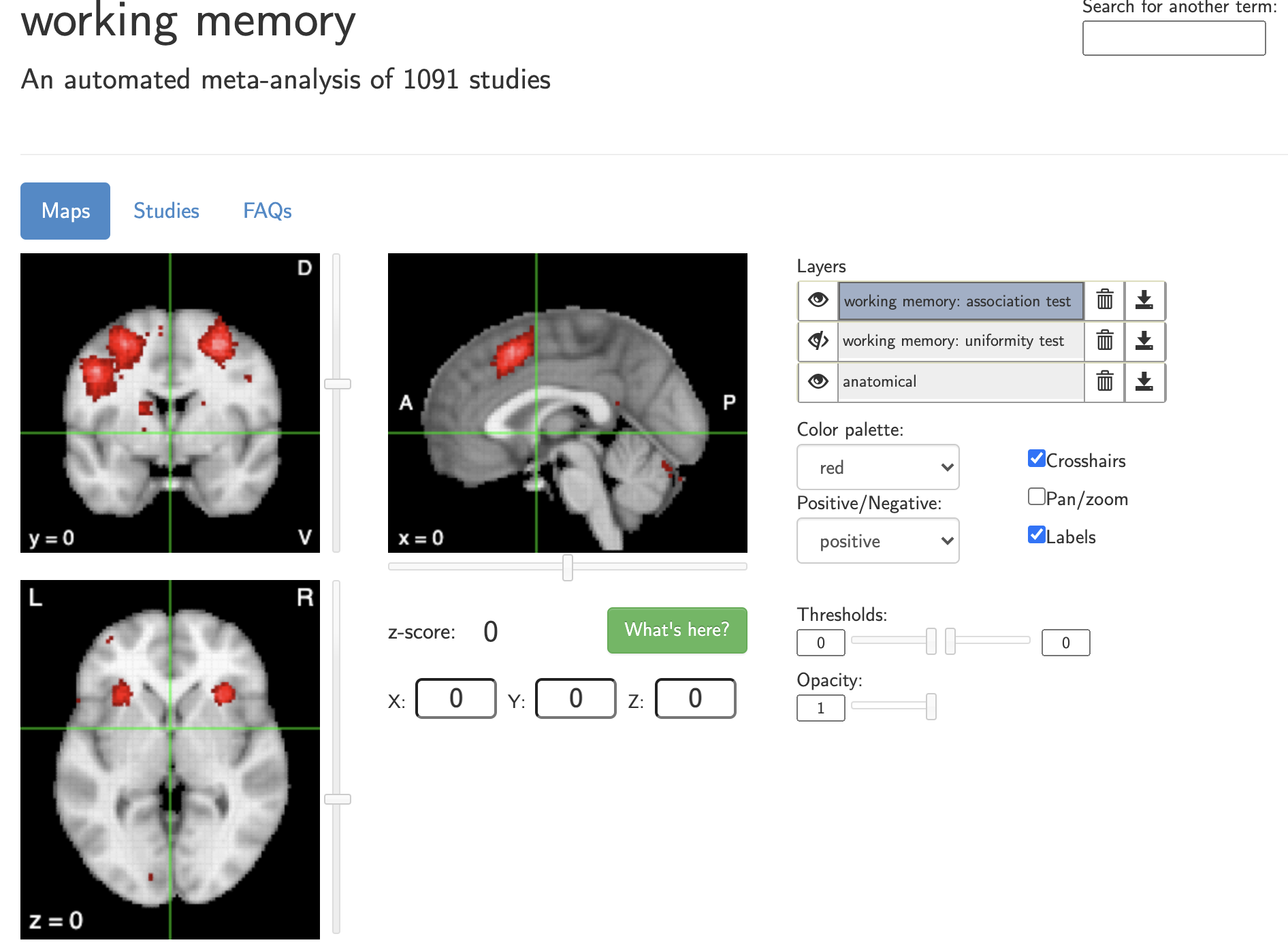


Decoding brain activity patterns using Neurosynth

Example of Neurosynth usage
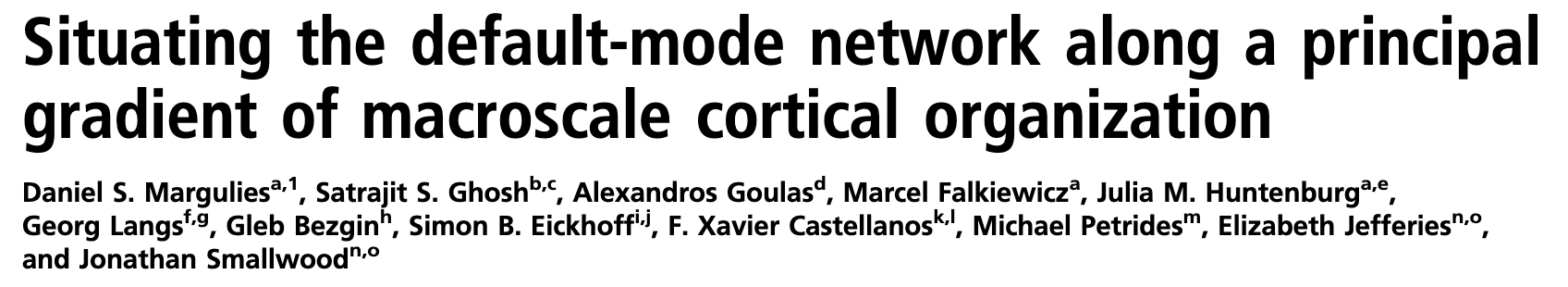

- Identified gradients of functional organization across the cortex
- Used Neurosynth to identify the most common terms associated with each gradient
Neurovault: Sharing neuroimaging results
- The results of most neuroimaging studies are images with statistical estimates at each voxel
- Neurovault.org is an open archive for these results

Gorgolewski et al., 2015, Frontiers in Neuroinformatics
- Collections
- A set of images (such as all images from a particular paper) can be uploaded as a collection
- Each collection receives a persistent identifier
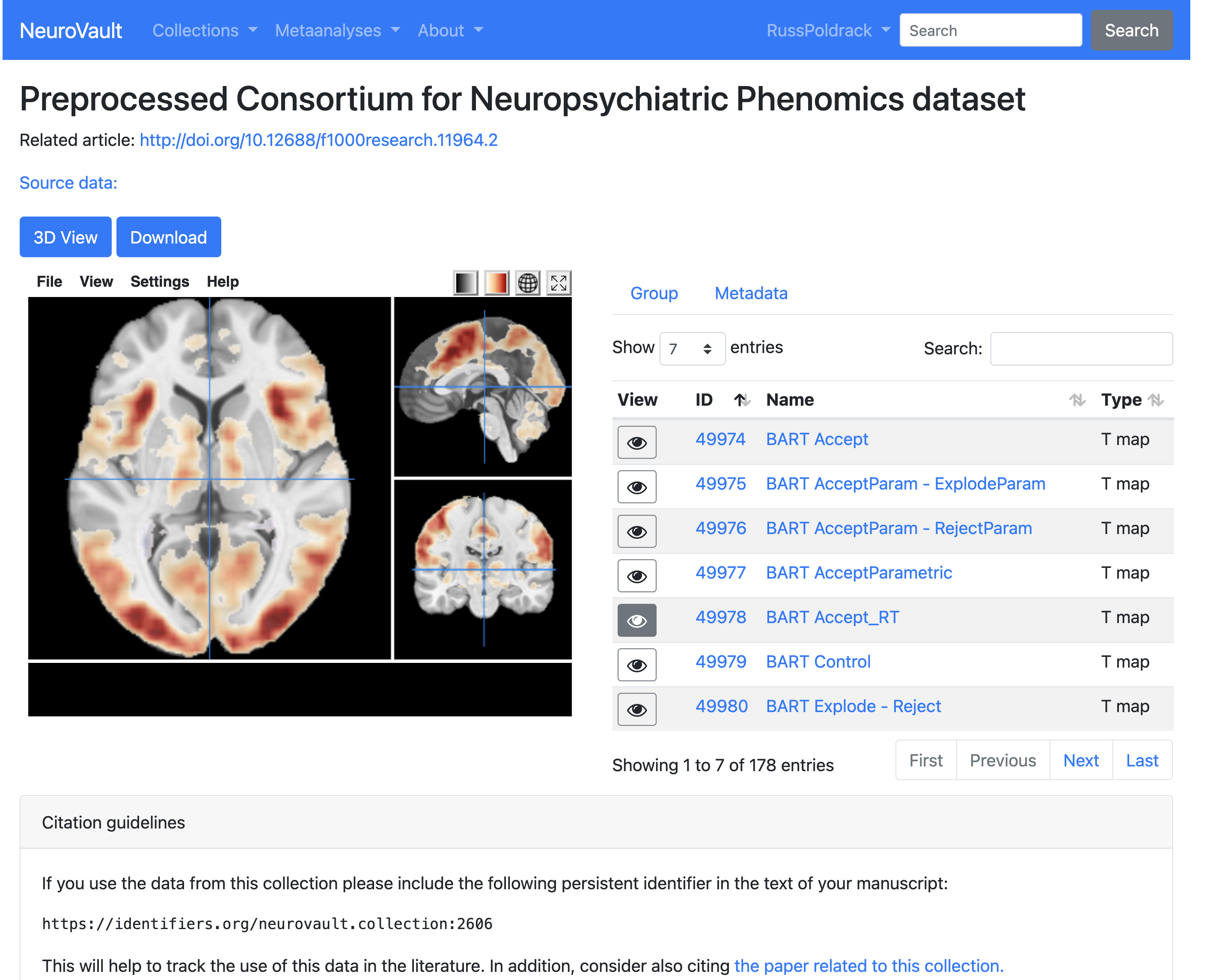
- Image browser
- Individual images can be browsed and downloaded
- A number of analysis tools can also be applied
- Each image also receives a persistent identifier
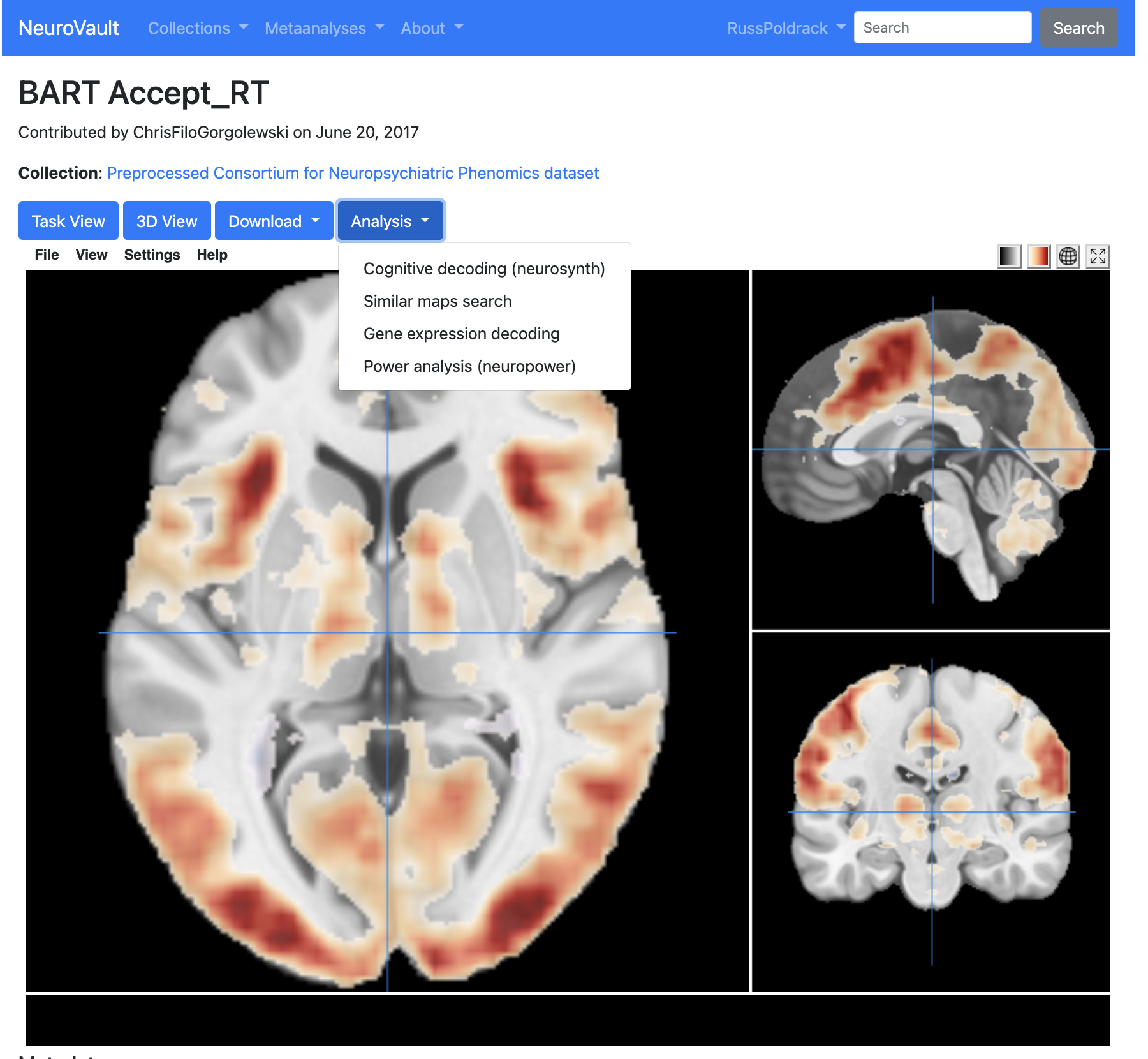
Example of Neurovault usage

OpenNeuro: Sharing raw and processed neuroimaging data

Simply sharing data is not sufficient
It must be shared in a way that makes it useful!
It’s easy to share data badly
Data Sharing and Management Snafu in 3 Short Acts

- I received the data, but when I opened it up it was in hexadecimal
- Yes, that is right
- I cannot read hexadecimal
- You asked for my data and I gave it to you. I have done what you asked.
…
- Is there a guide to the data anywhere?
- Yes, of course, it is the article that is published in Science.
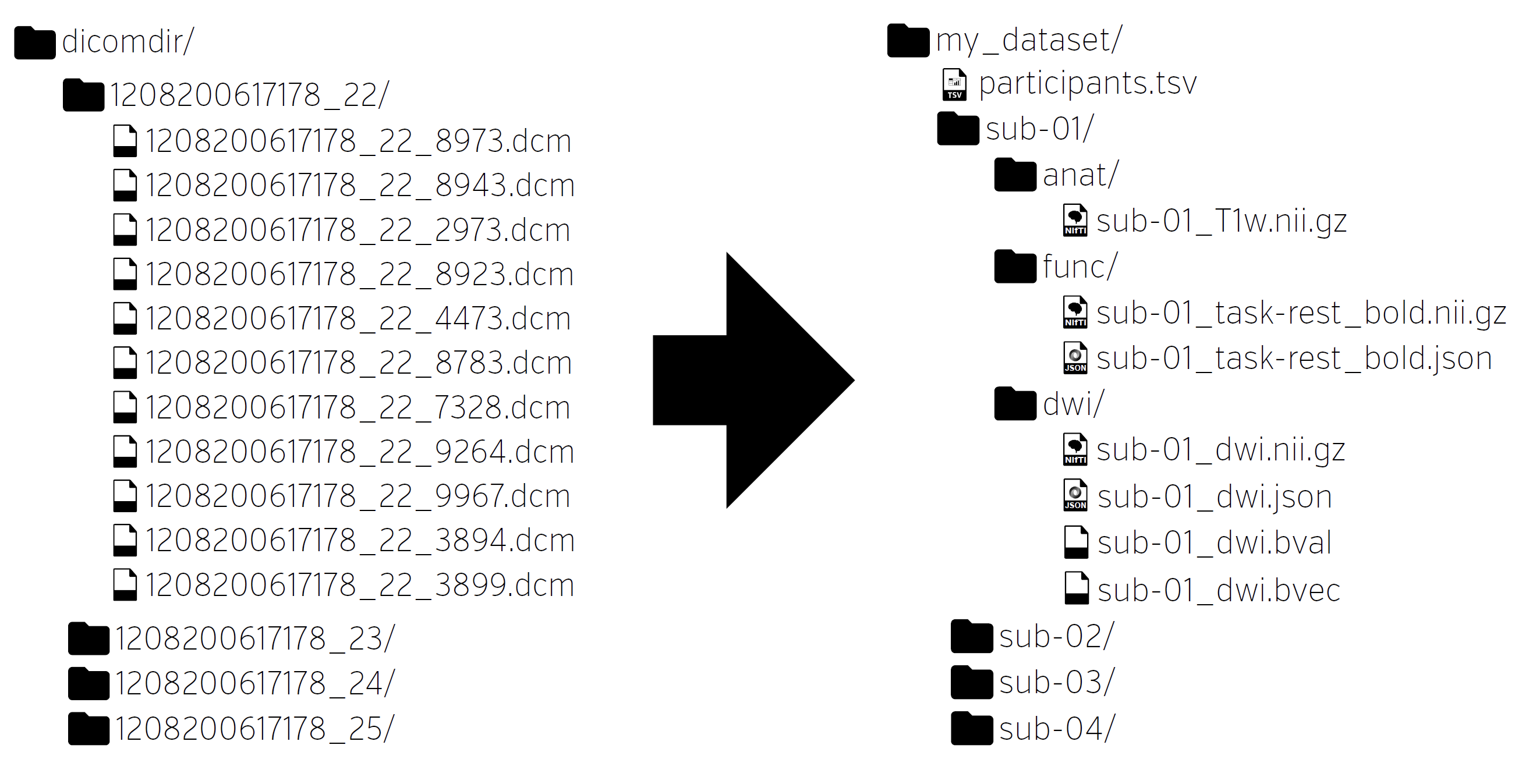
Brain Imaging Data Structure (BIDS)
- A community-based open standard for neuroimaging data
- A file organization standard
- A metadata standard

The development of BIDS
- January 2015
- Initial stakeholder meeting at Stanford (funded by INCF)
- Initiated development of a draft standard
- September 2015
- Draft standard posted to BIDS web site with 22 example datasets
- Solicited feedback from community
- June 2016
- Published paper
- September 2018
- BIDS-standard Github organization started
BIDS Principles
- Adoption is crucial
- Keep it as similar to existing practices as possible
- Don’t let technology override usability!
- Focus on engaging the community
- Keep it as similar to existing practices as possible
- Don’t reinvent the wheel
- Use existing standards when possible
- 80/20 rule
- Focus on the most common use cases
- Don’t let the perfect be the enemy of the good!
From DICOM to BIDS
The importance of automated validation

https://bids-standard.github.io/bids-validator/
BIDS Extensions
- BIDS was originally focused on structural/functional MRI data
- BIDS extension process allows extension of the standard through BIDS Extension Proposals (BEPS) initiated by the community
- Patterned after the Python Enhancement Proposal (PEP) process
| BEP # | Title |
|---|---|
| BEP001 | Quantitative MRI (qMRI) |
| BEP003 | Common Derivatives |
| BEP005 | Arterial Spin Labeling (ASL) |
| BEP006 | Electroencephalography (EEG) |
| BEP007 | Hierarchical Event Descriptor (HED) Tags |
| BEP008 | Magnetoencephalography (MEG) |
| BEP009 | Positron Emission Tomography (PET) |
| BEP010 | intracranial Electroencephalography (iEEG) |
| BEP018 | Genetic information |
| BEP030 | Near Infrared Spectroscopy (NIRS) |
| BEP031 | Microscopy |
The growing usage of BIDS: An example
- MRIQC Web API
- Crowdsourced database of MR QC metrics
- QC metrics from ~375K unique BOLD scans and ~280K T1w scans as of June 2022
- Publicly available: https://mriqc.nimh.nih.gov/


BIDS enables a growing open-source software ecosystem
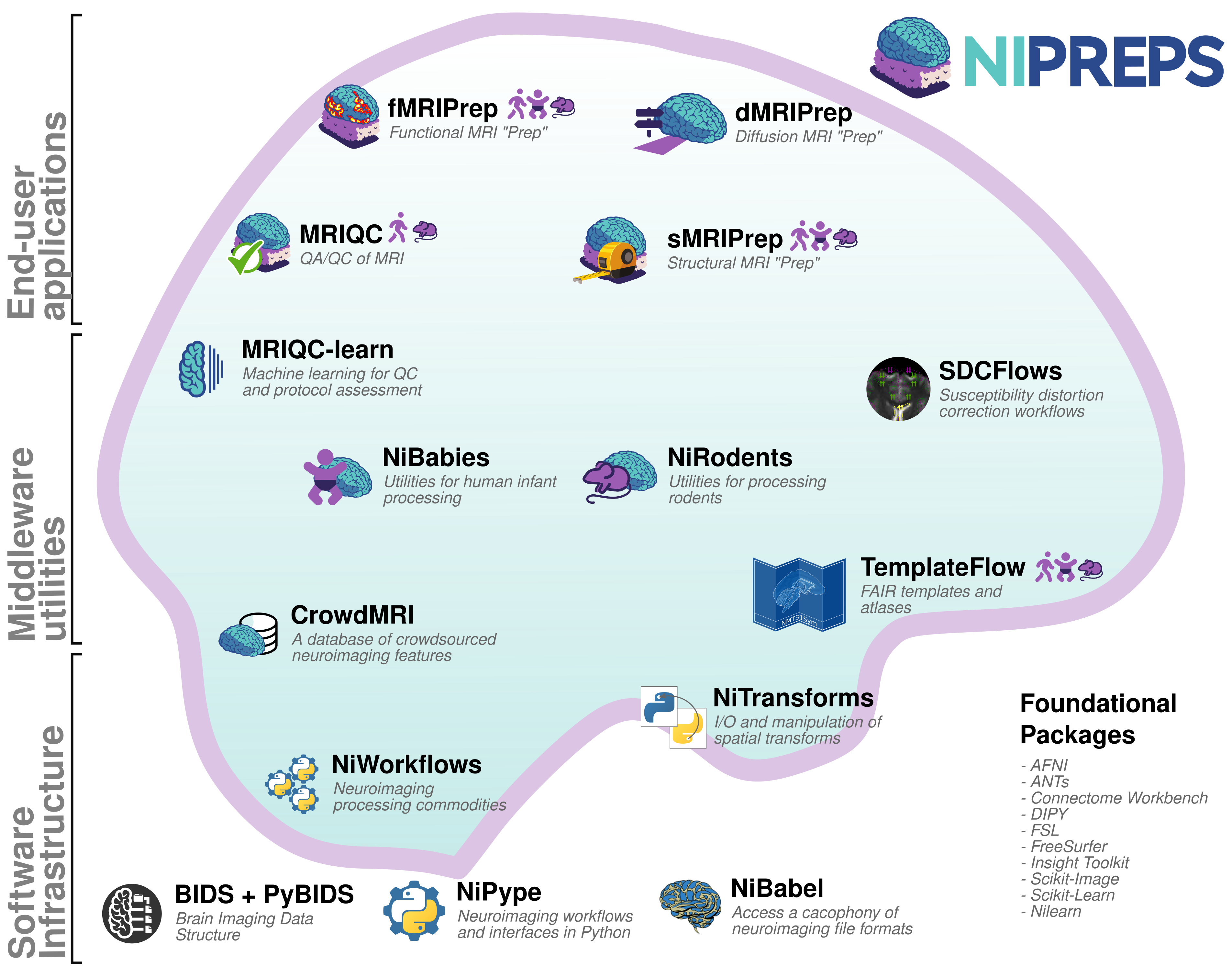
BIDS Apps


- Containerized applications that can be run on a BIDS dataset
- Containers provide ease of use as well as better reproducibility
fMRIprep: Robust preprocessing of fMRI data
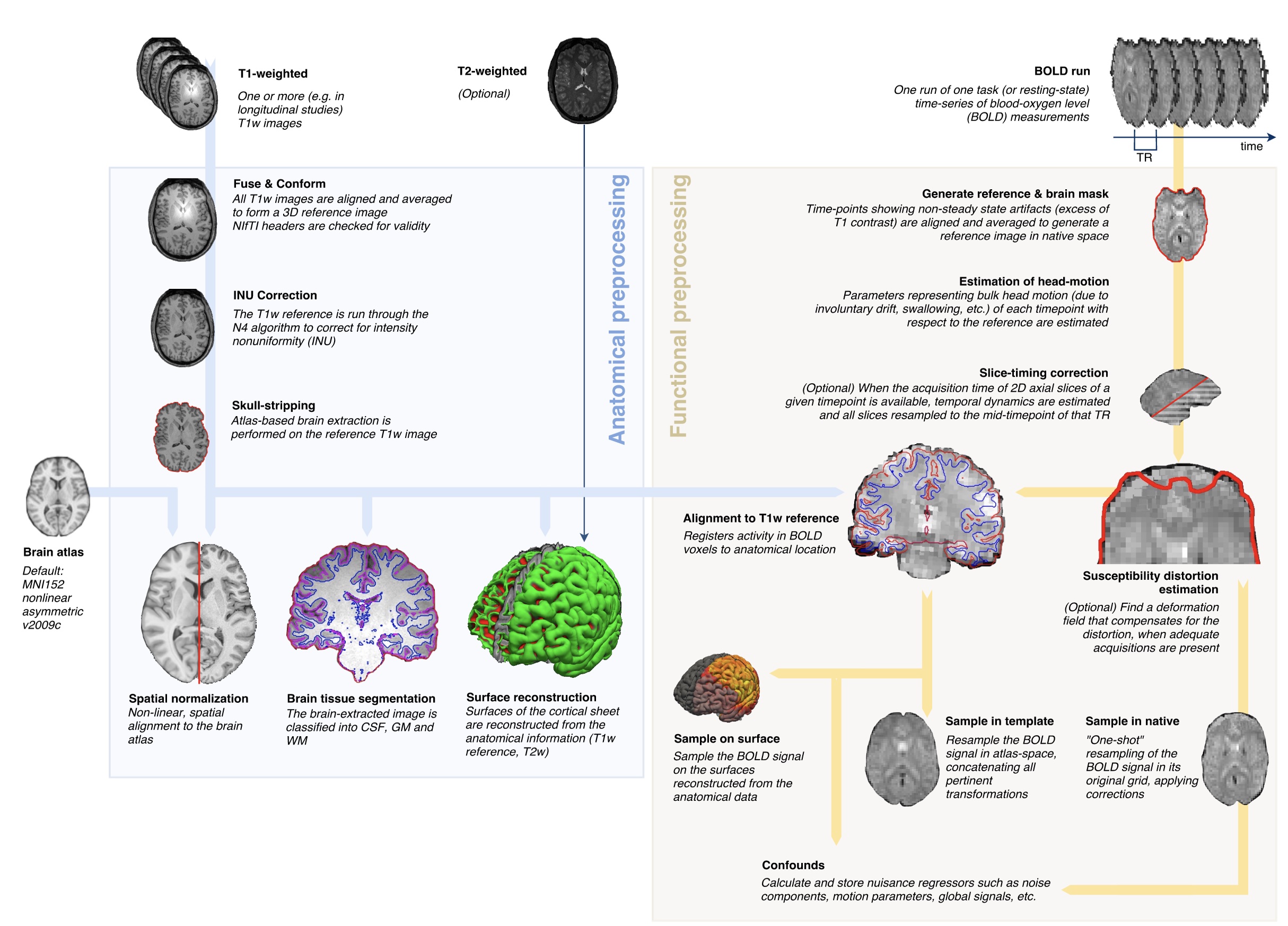
fmriprep.org; Esteban et al, 2019
MRIQC: MRI quality control for BIDS data
mriqc.org; Esteban et al, 2017
Tenmplateflow: FAIR Sharing of Neuroimaging Templates
- Templates and atlases are commonly used in neuroimaging
- There is a significant lack of clarity in the use of these templates
- There are numerous versions of the widely used “MNI template”
- Templateflow provides programmatic access to templates and mappings between them in a BIDS-like format
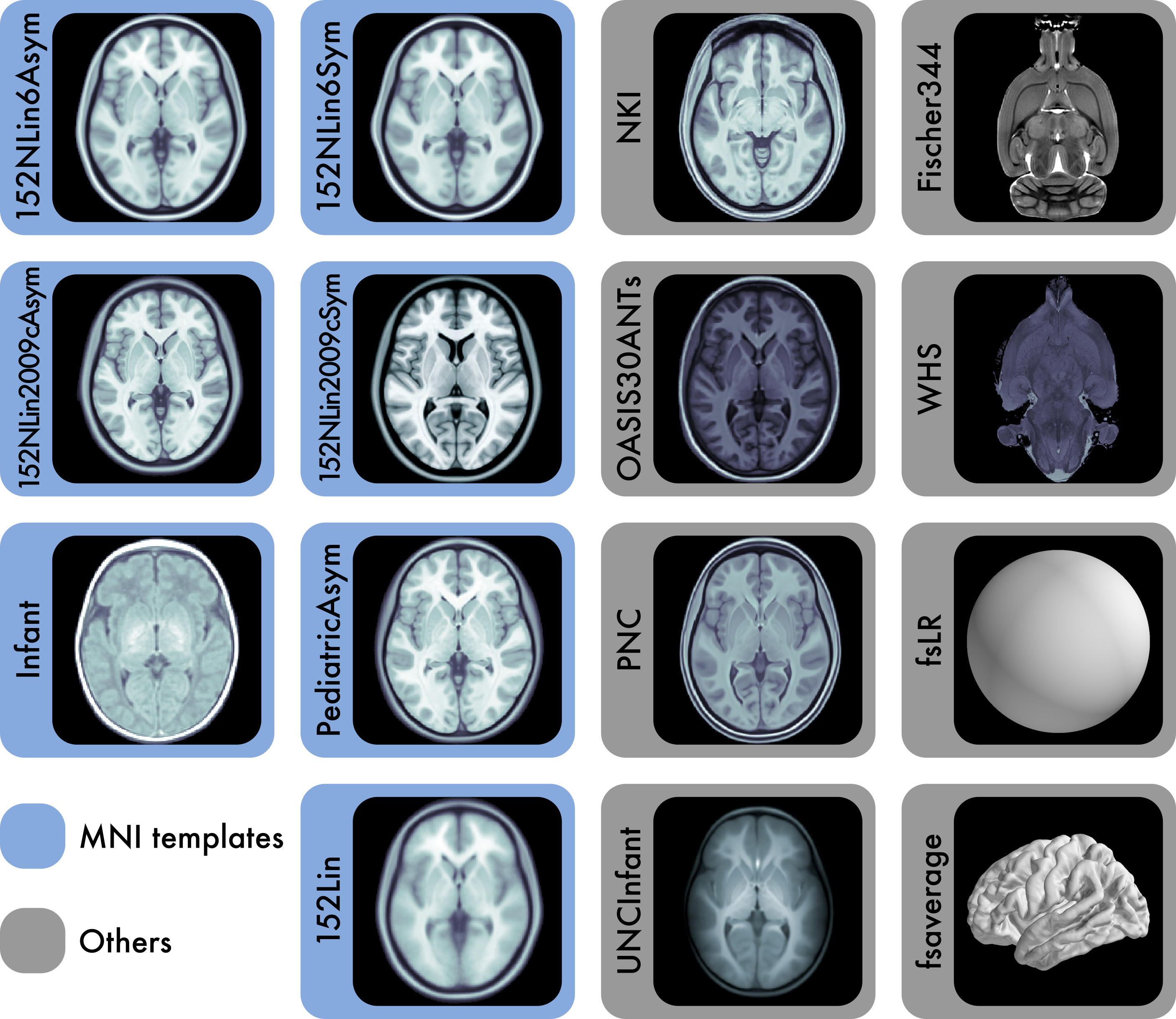
templateflow.org; Ciric et al., 2022
OpenNeuro: A BRAIN Initiative archive for BIDS data

- Supports sharing of any validated BIDS dataset
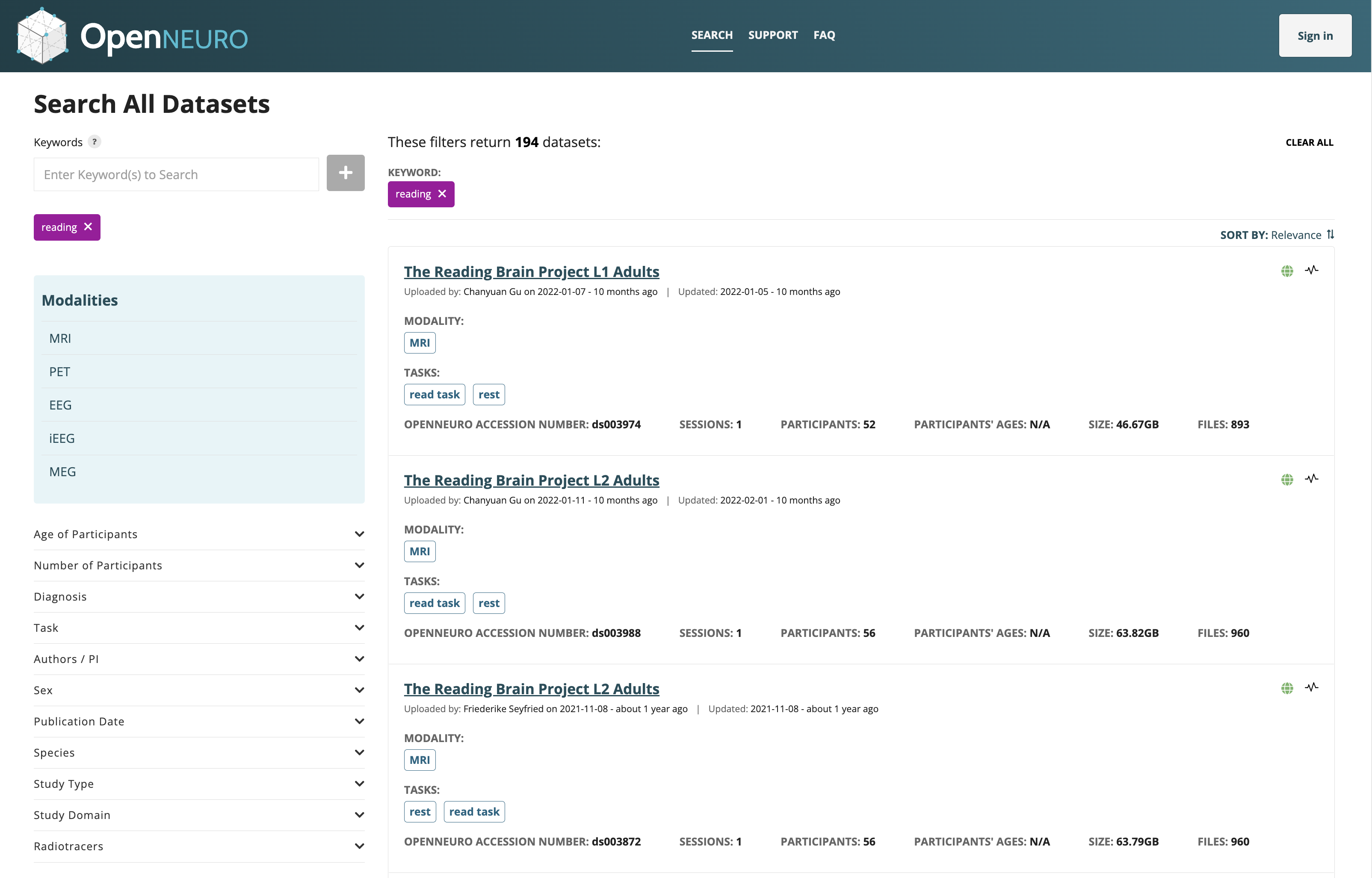
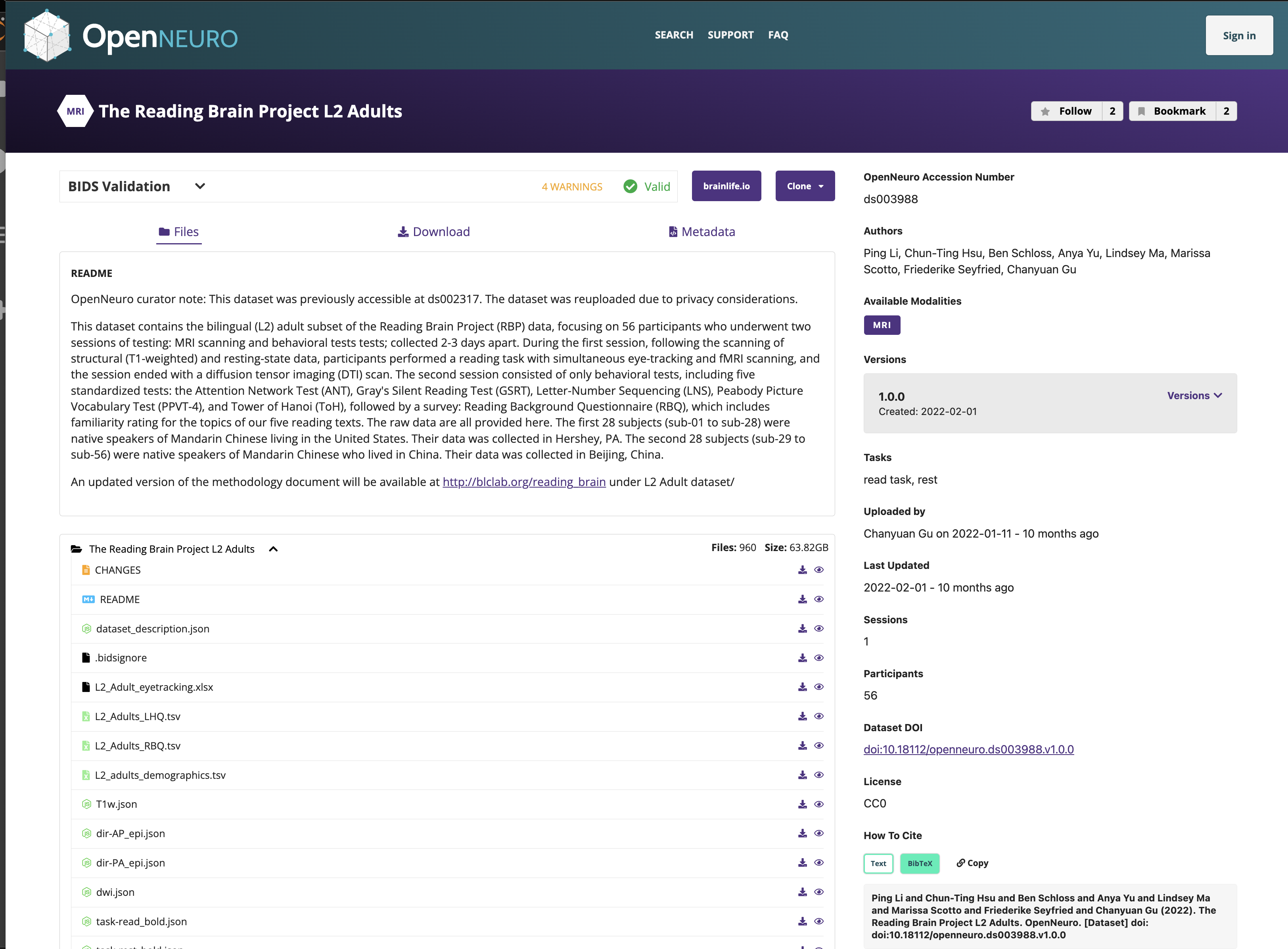
Each shared dataset is versioned and receives a persistent identifier (DOI)

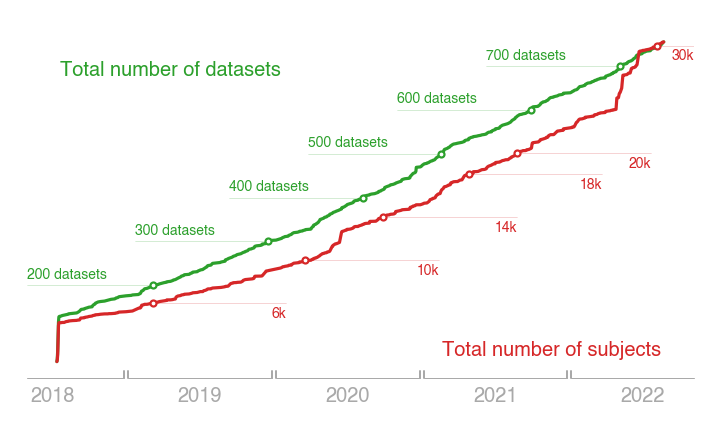
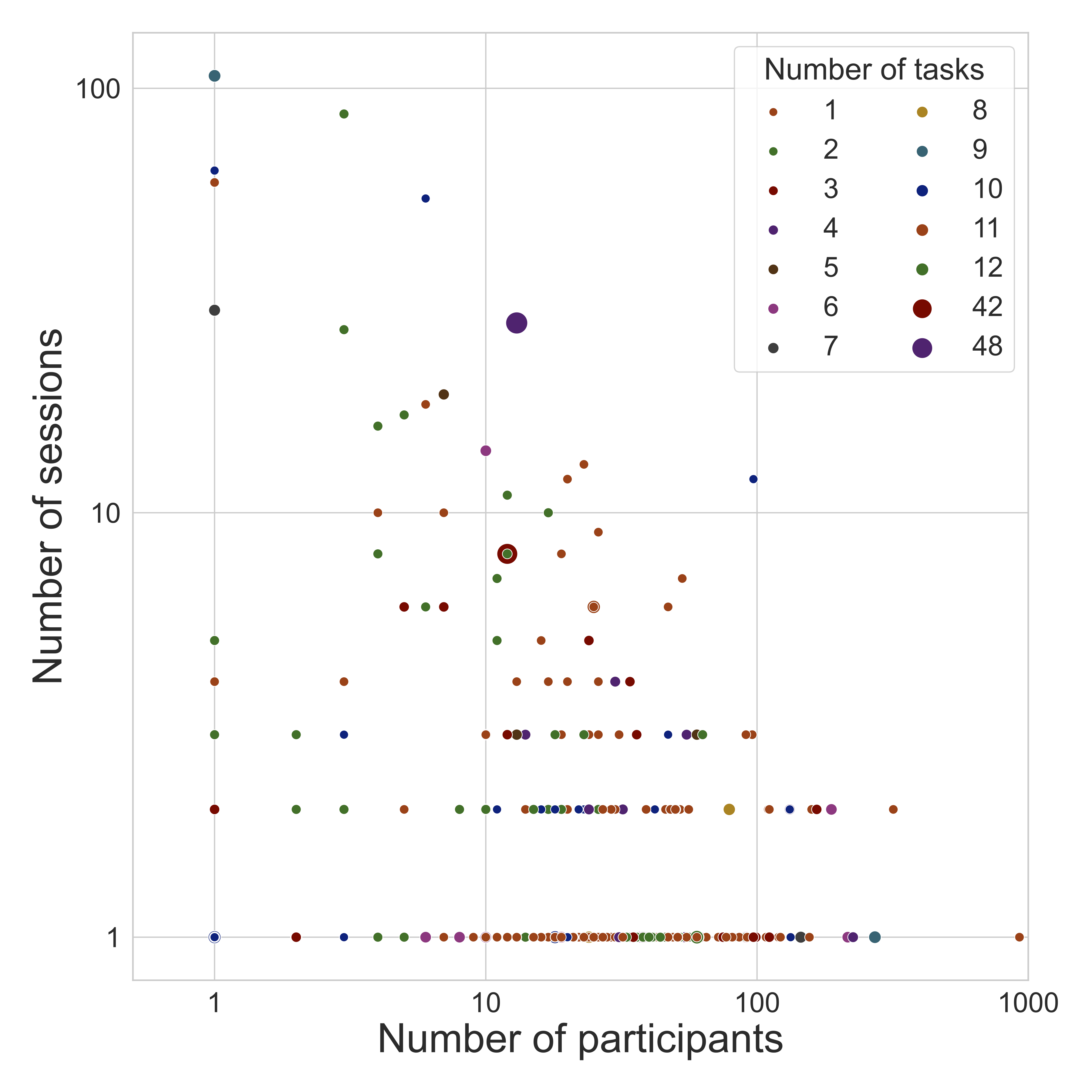
The growth of OpenNeuro
The diversity of OpenNeuro datasets
| Datatype | # |
|---|---|
| mri - anat | 597 |
| mri - func | 521 |
| eeg | 120 |
| mri - dwi | 67 |
| meg | 30 |
| ieeg | 17 |
| beh | 13 |
| pet | 11 |
| Species | # |
|---|---|
| Human | 676 |
| Mouse | 20 |
| Rat | 12 |
| NHP | 2 |
| phantoms | 1 |
| Juvenile pigs | 1 |
| Human, Mouse | 1 |
| Dog | 1 |
| Monkey | 1 |
| Sheep | 1 |
updated from Markiewicz et al, 2021, eLife
Scholarly reuse of OpenNeuro datasets
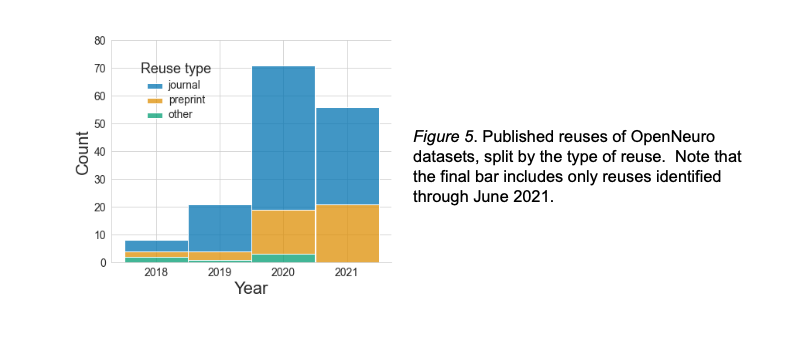
Markiewicz et al, 2021, eLife
Processing of OpenNeuro data
brainlife.io: processing of MRI data 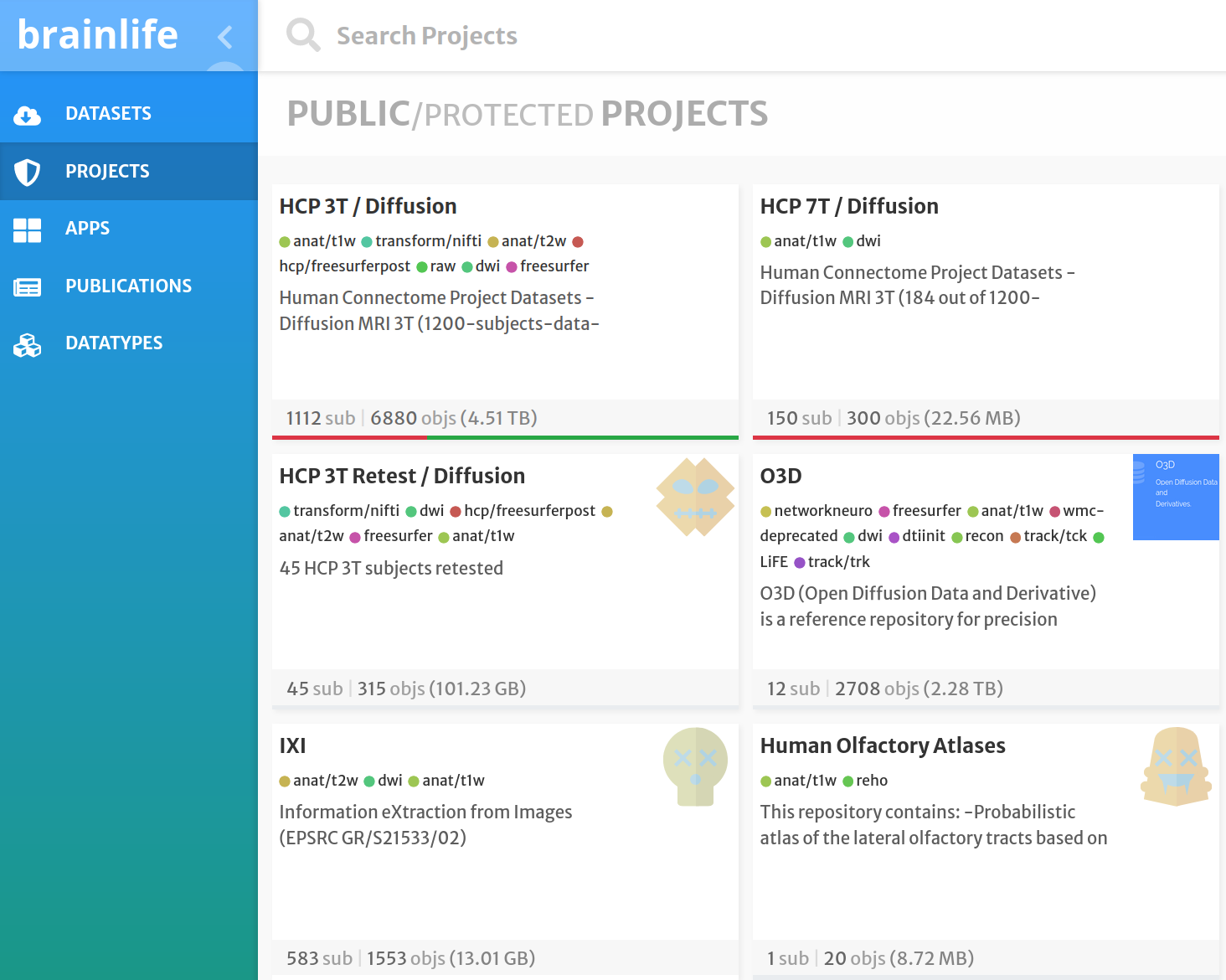
NEMAR.org: processing of EEG/MEG data 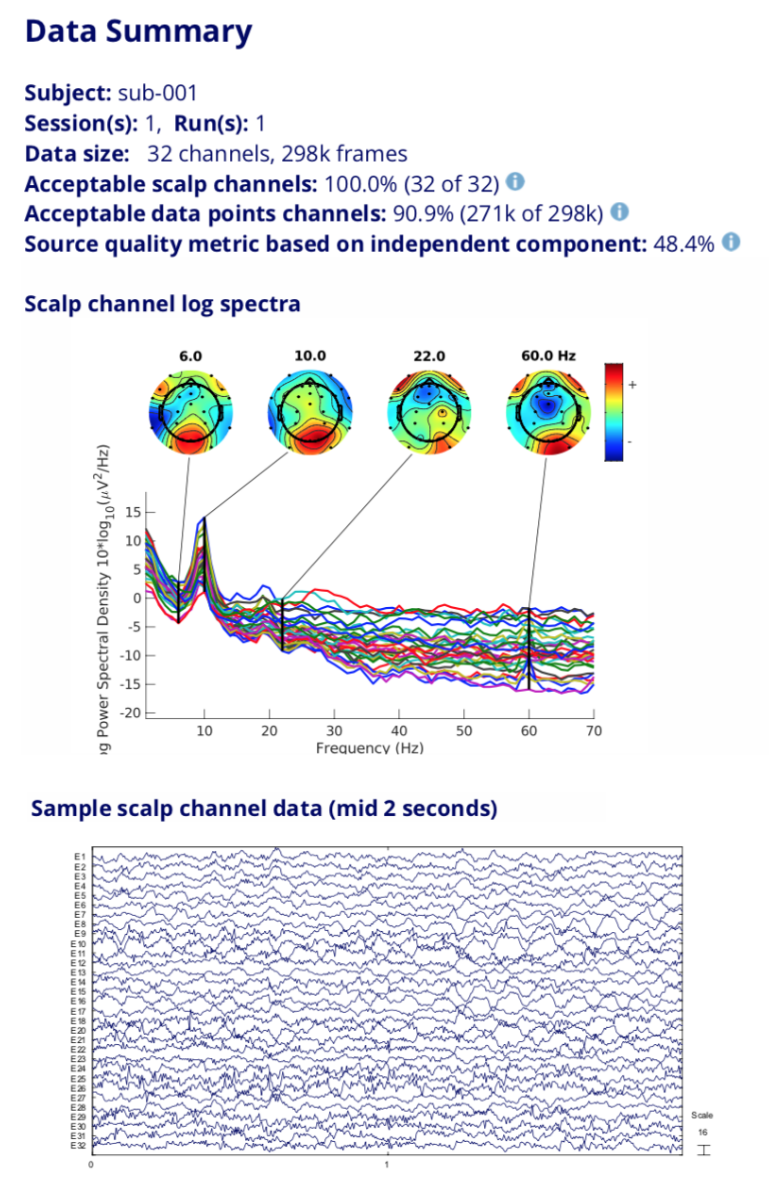
Example of OpenNeuro reuse
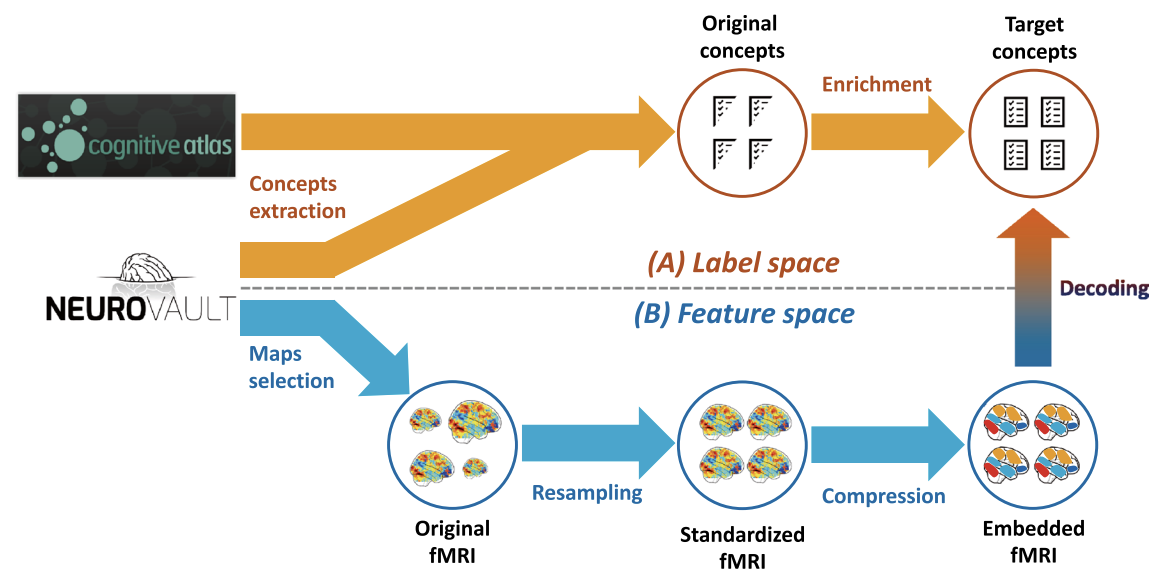
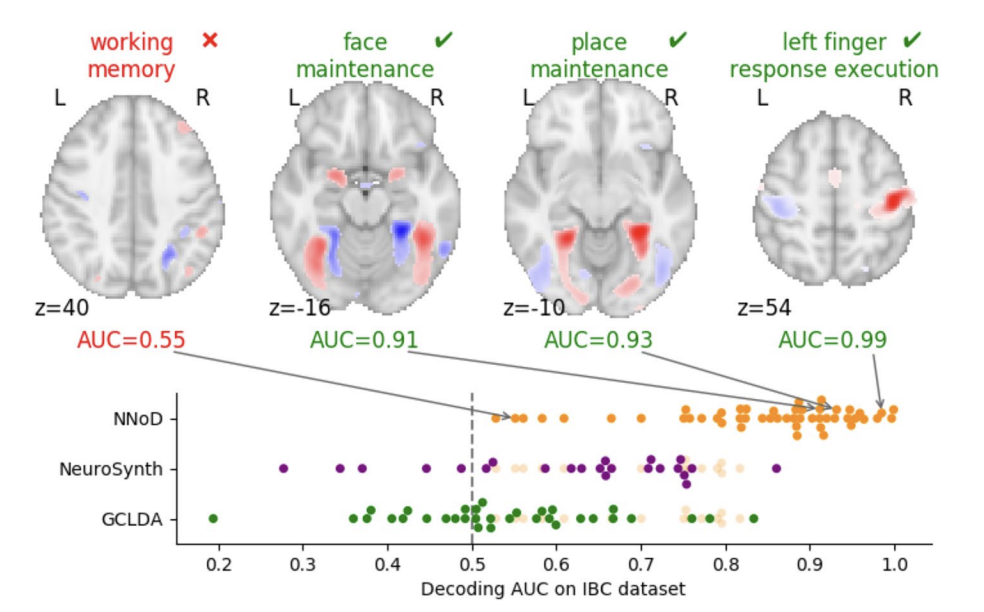
- A challenge for decoding brain activity from fMRI data is that most datasets are very small
- We used OpenNeuro to train a “foundation model”
- A pre-trained model that can be used as a starting point for decoding models on smaller datasets
- We pre-train models on broad fMRI data from OpenNeuro: 11,980 experimental runs from 1,726 individuals across 34 datasets.
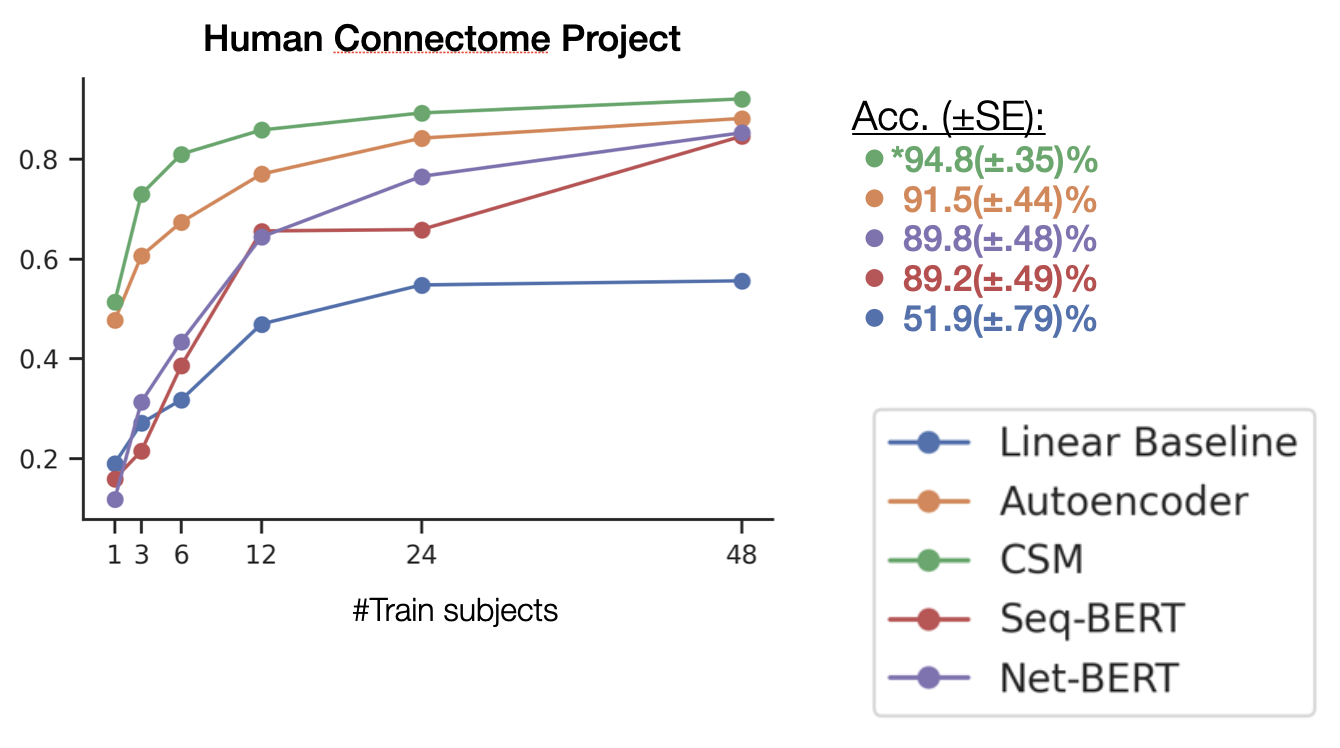
- This approach substantially increased decoding performance vs. a baseline model
Thomas, Ré, & Poldrack, 2022, NeurIPS
Challenges to open sharing
- All OpenNeuro MRI datasets must be defaced
- To reduce risk of reidentification
- There is increasing risk that subjects might be reidentified even after defacing using advanced face recognition systems + face imputation tools (Schwartz et al., 2021)
- If the risk continues to rise, it may become necessary to move away from open sharing
- This would be a huge loss for researchers, research participants, and the world
- We have proposed regulatory changes to protect subjects from misuse of neuroscience information in the US context (Jwa & Poldrack, 2022, J. Law & Biosciences)

Keys to success in neuroimaging data sharing
- Data are digital end-to-end
- Minimizes manual steps in the process
- Standardized file formats and data standards
- Makes data immediately usable by anyone
- Reduces burden of curation and preparation
- Demonstrated scientific utility
- Numerous success stories
Lessons learned
- Community buy-in is essential
- Mandates put in place before the community is ready can backfire
- Unless they have overwhelmingly powerful advocates, as in genomics
- Important that sharing advocates are members of community and eat their own dog food
- Mandates put in place before the community is ready can backfire
Lessons learned
- Keep it simple and as close to standard practice as possible
- Overengineered solutions have generally failed
- If there are more than 2 acronyms…
Lessons learned
- Don’t let the perfect be the enemy of the good
- 20% of the effort will cover 80% of the datasets - focus on these!
- There is a long tail of edge cases with loud advocates

Vilfredo Pareto
Conclusions
- The field of neuroimaging has built an model ecosystem for open science and data sharing
- Infrastructure is critical to ease friction
- Community engagement has been key to adoption
- Need to keep the tools as close as possible to current practice
The Poldrack Lab

Funding




OpenNeuro Team






Collaborators




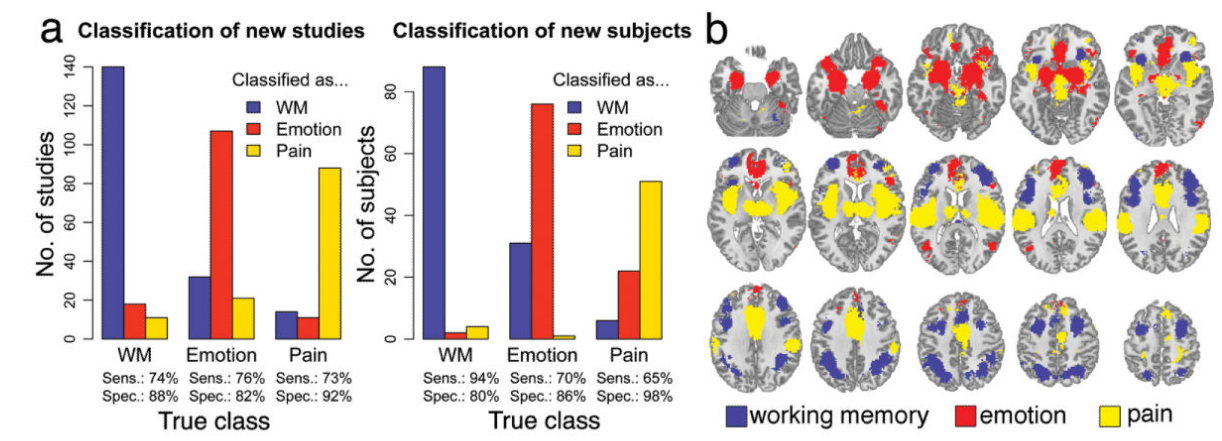
Meta-analytic decoding using Neurosynth
- Given 2+ terms, can determine which is most likely given the data
- Naive Bayes classifier: assumes that all features (voxels) are independent; selects the most probable class
- Can apply this to any activation map—studies, individual subjects, etc.

Yarkoni et al, 2011, Nature Methods
- Cross-validated classification of all studies in database
- Select 25 high-frequency terms
- Pairwise classification: how well can we distinguish between the presence of each pair of terms?
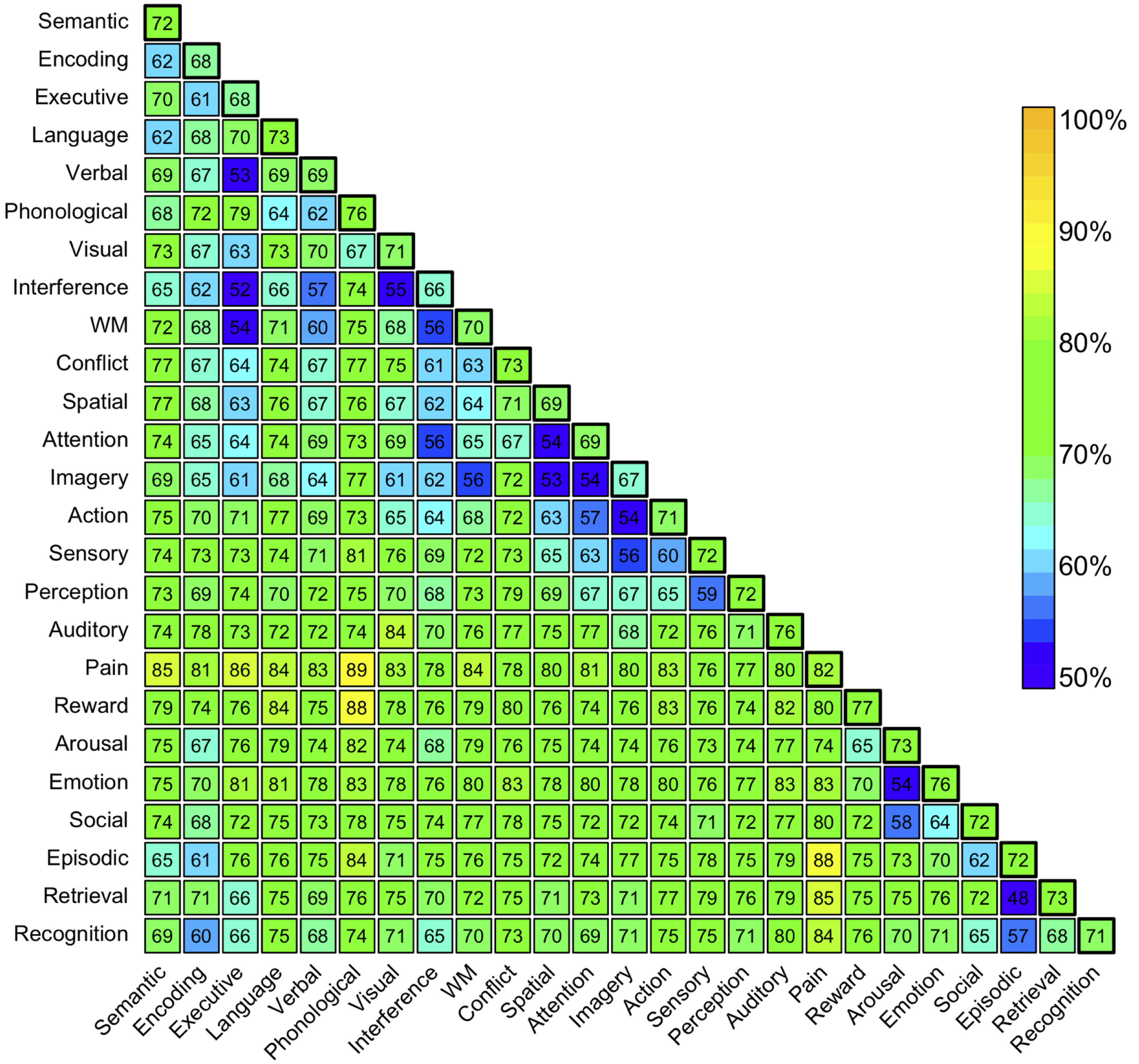
Yarkoni et al, 2011, Nature Methods
https://poldrack.github.io/talks-OpenScienceEcosystem/